The SARS-CoV-2 spike protein induces lung cancer migration and invasion in a TLR2-dependent manner
Mi-Jeong Kim, Ji Young Kim and Ji Hye Shin has contributed equally to this work
Abbreviations
-
- SARS-CoV-2
-
- severe acute respiratory syndrome coronavirus 2
-
- TLR
-
- toll-like receptor
-
- NSCLC
-
- non-small cell lung cancer
-
- ACE2
-
- angiotensin-converting enzyme 2
-
- TMPRSS2
-
- transmembrane protease serine subtype 2
-
- GSEA
-
- gene set enrichment analysis
-
- Pam3CSK4
-
- tripalmitoyl-S-glycero-Cys-(Lys) 4
-
- FSL-1
-
- fibroblast stimulating lipopeptide 1
-
- NF-κB
-
- nuclear factor kappa-light-chain-enhancer of activated B cells
-
- IKK
-
- inhibitor of nuclear factor-κB kinase
-
- ERK
-
- extracellular signal-regulated kinase
-
- CRISPR-Cas9
-
- clustered regularly interspaced short palindromic repeats and CRISPR-associated protein 9
Dear editor,
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection can lead to severe outcomes in patients with cancer [1]. It has been reported that patients with lung cancers disproportionately manifest severe COVID-19 with a high rate of hospitalization and death [2]. Notably, the SARS-CoV-2 Spike (S) protein can induce hyper-inflammation in both epithelial cells and macrophages through toll-like receptor (TLR)1/TLR2 or TLR2/6-dependent nuclear factor-kappaB (NF-κB) pathway [3]. However, molecular and cellular evidence on whether the SARS-CoV-2 virus affects the severity of lung cancer patients through TLR1/2 or TLR2/6 signaling remains unclear.
To obtain insight into the association of SARS-CoV-2 susceptibility and severity in lung cancer, we utilized microarray data of 42 non-small cell lung cancer (NSCLC) patients and analyzed different magnitude differences (∆Mag) of angiotensin converting enzyme 2 (ACE2) and TLR2 expression (termed ∆ACE2 and ∆TLR2), which are associated with SARS-CoV-2 infection and hyper-immune response [1-3], between lung tumor tissues and matched lung normal tissues (Supplementary Table S1). Based on ∆ACE2 and ∆TLR2, we selected 11 ACE2upTLR2up lung tumor tissues and 11 ACE2downTLR2down lung tumor tissues (Supplementary Figure S1A) and performed gene set enrichment analysis (GSEA) (https://www.gsea-msigdb.org) to determine whether expression levels of ACE2 and TLR2 were statistically associated with gene sets related to cancer, SARS-CoV-2 infection, and TLR signaling. Ten gene sets of cancer modules were significantly enriched in ACE2upTLR2up lung tumor tissues versus ACE2downTLR2down lung tumor tissues (Supplementary Figure S1B). Additionally, gene sets related to SARS-CoV-2 infection (Supplementary Figure S2A) or the TLR signaling pathway (Supplementary Figure S2B) were significantly enriched in ACE2upTLR2up lung tumor tissues. To verify these results in detail, we further analyzed ∆Mag of transmembrane protease serine subtype 2 (TMPRSS2), which contributes to virulence and pathogenesis of SARS-CoV-2 virus along with ACE2 [1, 2], TLR1, and TLR6, which are functionally formed as a TLR1/2 or TLR2/6 heterodimer [3], between lung tumor tissues and matched lung normal tissues (Figure 1A, Supplementary Table S1). Based on ∆ACE2, ∆TMPRSS2, ∆TLR1, ∆TLR2, and ∆TLR6, we further selected 4 ACE2upTMPRSS2upTLR1upTLR2upTLR6up lung tumor tissues and 7 ACE2downTMPRSS2downTLR1downTLR2downTLR6down lung tumor tissues (Figure 1A) and performed GSEA. Nine gene sets of cancer modules regulating cancer progression and development and 2 gene sets of cancer modules involving anti-apoptosis or inflammatory response were enriched in ACE2upTMPRSS2upTLR1upTLR2upTLR6up lung tumor tissues (Figure 1B, Supplementary Figure S3).
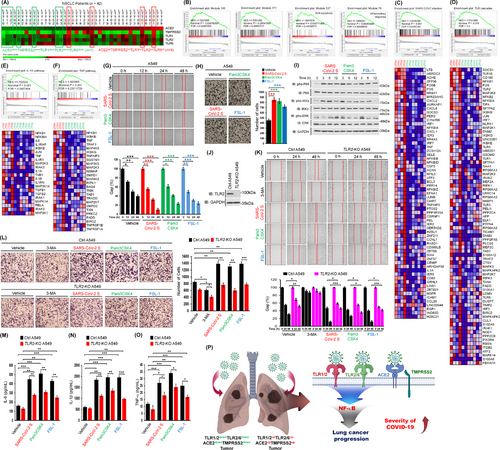
SARS-CoV-2 spike protein induces lung cancer migration and invasion in a TLR2-dependent manner. (A) 42 lung tumor tissues of NSCLC patients were listed according to the ∆Mag of ACE2, TMPRSS2, TLR1, TLR2, and TLR6 expression in lung tumor tissues (LTTs) versus matched lung normal tissues (mLNTs). Eleven lung tumor tissues, four ACE2upTMPRSS2upTLR1upTLR2upTLR6up lung tumor tissues and seven ACE2downTMPRSS2downTLR1downTLR2downTLR6down lung tumor tissues, were selected for GSEA. The color scale indicates ∆Mag value. (B) GSEA was performed in four ACE2upTMPRSS2upTLR1upTLR2upTLR6up lung tumor tissues versus seven ACE2downTMPRSS2downTLR1downTLR2downTLR6down lung tumor tissues indicated in (A). Gene sets for four cancer modules are presented. (C-F) Gene sets for ACE2 expressing cells with SARS-CoV-2 infection (C), TLR cascades (D), IL-1R pathway (E), and TNF pathway (F) are presented along with heat maps showing differential gene expression patterns between four ACE2upTMPRSS2upTLR1upTLR2upTLR6up lung tumor tissues and seven ACE2downTMPRSS2downTLR1downTLR2downTLR6down lung tumor tissues. NES, nominal P-value, and FDR q-values are indicated in the inner panel. (G) A549 lung cancer cells were treated with vehicle, SARS-CoV-2 S protein, Pam3CSK4, or FSL-1 (up). The residual gap between migrating cells from the opposing wound edge is expressed as a percentage of the initial scraped area (± SD, n = 3 different plates) (down). (H) A549 lung cancer cells were treated with vehicle, SARS-CoV-2 S protein, Pam3CSK4, or FSL-1 for 24 h (left). The number of migrating cells was counted. Results are presented as mean ± SD of three independent experiments (right). (I) A549 lung cancer cells were stimulated with SARS-CoV-2 S protein, Pam3CSK4, or FSL-1. Phosphorylation levels of P65, IKKs, and ERK were measured by Western blotting. (J) TLR2-knockout (TLR2-KO) A549 cells were generated using CRISPR-Cas9 gene-editing method. TLR2 expression was verified by western blotting with anti-TLR2 and anti-GAPDH (control). (K) Ctrl A549 and TLR2-KO A549 cells were treated with vehicle, 3-MA, SARS-CoV-2 S protein, Pam3CSK4, or FSL-1 for different time periods (up). The residual gap between migrating cells from the opposing wound edge is expressed as a percentage of the initial scraped area (± SD, n = 3 different plates) (down). (L) Ctrl A549 and TLR2-KO A549 cells were treated with vehicle, 3-MA, SARS-CoV-2 S protein, Pam3CSK4, or FSL-1 for 24 h (left). The number of migrating cells was counted. Results are presented as mean ± SD of three independent experiments (right). (M-O) Ctrl A549 and TLR2-KO A549 cells were treated with vehicle, SARS-CoV-2 S protein, Pam3CSK4, or FSL-1 for 12 h. Production levels of IL-6 (M), IL-1β (N), and TNF-α (O) in culture supernatant were measured by ELISA. Results are presented as mean ± SD of three independent experiments. (P) A schematic model of how the SARS-CoV-2 virus critically affects the susceptibility to SARS-CoV-2 infection and the severity of SARS-CoV-2 infection in lung cancer patients with up-regulation of ACE2, TMPRSS2, TLR1, TLR2, and TLR6. *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; TLR, toll-like receptor; NSCLC, non-small cell lung cancer; ∆Mag; magnitude difference; ACE2, angiotensin-converting enzyme 2; TMPRSS2, transmembrane protease serine subtype 2; LTTs, lung tumor tissues; mLNTs, matched lung normal tissues; GSEA, gene set enrichment analysis; IL-1R, interleukin-1 receptor; TNF, tumor necrosis factor; Pam3CSK4, tripalmitoyl-S-glycero-Cys-(Lys) 4; FSL-1, fibroblast stimulating lipopeptide 1; TLR2-KO, TLR2-knockout; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; ELISA, enzyme-linked immunosorbent assay; IKK, inhibitor of nuclear factor-κB kinase; ERK, extracellular signal-regulated kinase; CRISPR-Cas9, clustered regularly interspaced short palindromic repeats and CRISPR-associated protein 9; IL-6, interleukin 6.
Remarkably, gene sets upregulated in SARS-CoV-2 infection and innate signaling pathways, including TLR cascades and the nucleotide-binding and oligomerization domain (NOD)-like receptor pathway, were significantly enriched in ACE2upTMPRSS2upTLR1upTLR2upTLR6up lung tumor tissues versus ACE2downTMPRSS2downTLR1downTLR2downTLR6down lung tumor tissues (Figure 1C-D, Supplementary Figure S4A), along with their downstream signaling targets and pathways, such as the NF-κB pathway, NF-κB1 targets, tumor necrosis factor (TNF) signaling via NF-κB, interferon-α/β signaling, the interleukin-1 receptor (IL-1R) pathway, and the TNF pathway (Figure 1E-F, Supplementary Figure S4B-E). SARS-CoV-2 virus can infect human cells and induce inflammatory cytokines and chemokines, including IL-6, IL-1β, TNF-α, C-X-C motif chemokine ligand 1 (CXCL1), CXCL2, and C-C motif chemokine ligand 2 (CCL2), via TLR2-dependent activation of the NF-κB pathway [3-6]. Therefore, we further assessed whether expression levels of ACE2, TMPRSS2, TLR1, TLR2, and TLR6 were associated with gene sets related to inflammatory cytokines and chemokines. A gene set related to cytokine receptor interaction was highly enriched in ACE2upTMPRSS2upTLR1upTLR2upTLR6up lung tumor tissues versus ACE2downTMPRSS2downTLR1downTLR2downTLR6down lung tumor tissues (Supplementary Figure S5A). Gene sets related to signal transduction through IL-1R, tumor necrosis factor receptor 2 (TNFR2) pathway, TNF targets, and cytokine pathways were enriched in ACE2upTMPRSS2upTLR1upTLR2upTLR6up lung tumor tissues versus ACE2downTMPRSS2downTLR1downTLR2downTLR6down lung tumor tissues (Supplementary Figure S5B-G). Additionally, gene sets of the IL-8-C-X-C motif chemokine receptor 1 (CXCR1), IL-8-CXCR2, and cytokine- and chemokine-mediated signals were enriched in ACE2upTMPRSS2upTLR1upTLR2upTLR6up lung tumor tissues (Supplementary Figure S5H-M). These results suggest that upregulation of ACE2, TMPRSS2, TLR1, TLR2, and TLR6 in lung cancer tissues are associated with gene sets related to cancer progression, SARS-CoV-2 infection, and inflammatory responses.
Given that the SARS-CoV-2 S protein can induce inflammation via TLR1/2- or TLR2/6-dependent activation of the NF-κB pathway [3], we investigated whether the SARS-CoV-2 S protein could induce lung cancer migration, invasion, colony formation, and cell proliferation via TLRs. Upon treatment with the SARS-CoV-2 S protein, Pam3CSK4 (an agonist of TLR1/2), or FSL-1 (an agonist of TLR2/6), migration and invasion abilities of A549 and H1299 lung cancer cells were significantly enhanced compared to those upon treatment with vehicle control (Figure 1G-H, Supplementary Figure S6). Moreover, colony-forming and cell proliferation assay revealed significant increases in response to the SARS-CoV-2 S protein, Pam3CSK4, or FSL-1 (Supplementary Figure S7). Notably, stimulation of the SARS-CoV-2 S protein, Pam3CSK4, or FSL-1 induced increases in phosphorylation of P65, inhibitor of nuclear factor-κB kinases (IKKs), and extracellular signal-regulated kinase (ERK) compared to treatment with vehicle in A549 cells (Figure 1I, Supplementary Figure S8A-C), accompanied with increases in activities of cytokines such as IL-6, IL-1β, TNF-α, and NF-κB (Supplementary Figure S8D-G). Consistent with data of A549 cells, H1299 cells treated with the SARS-CoV-2 S protein, Pam3CSK4, or FSL-1 showed elevated levels of phosphorylated P65, IKKs, and ERK (Supplementary Figure S9A-D) and production of IL-6, IL-1β, and TNF-α (Supplementary Figure S9E-G). These results suggest that the SARS-CoV-2 S protein can induce lung cancer progression by activating the NF-κB pathway via TLR1/2 and TLR2/6.
To directly verify the functional role of the SARS-CoV-2 S protein in lung cancer progression through TLR1/2 and TLR2/6 stimulation, we generated TLR2-knockout (KO) A549 and H1299 cells using CRISPR-Cas9 gene-editing method [6-9] (Figure 1J, Supplementary Figure S10) and performed cancer progression assay as previously described [7-9]. Consistently, treatment with the SARS-CoV-2 S protein, Pam3CSK4, or FSL-1 enhanced cancer migration and invasion of control (Ctrl) A549 cells as compared to treatment with vehicle control, whereas TLR2-KO A549 cells showed markedly attenuated cancer migration and invasion (Figure 1K-L). On the other hand, 3-methyladenine (3-MA), an inhibitor of phosphoinositide-3-kinase (PI3K), inhibited cell migration and invasion of Ctrl A549 and TLR2-KO A549 cells (Figure 1K-L). Furthermore, TLR2-KO A549 cells treated with the SARS-CoV-2 S protein, Pam3CSK4, or FSL-1 showed significantly reduced production of IL-6, IL-1β, and TNF-α as compared to Ctrl A549 cells (Figure 1M-O). We observed similar results of cell migration and invasion of TLR2-KO H1299 cells (Supplementary Figure S11). As depicted in Figure 1P, these results suggest that the SARS-CoV-2 S protein can directly induce lung cancer progression, including migration, invasion, colony formation, and proliferation, in a TLR2-dependent manner.
In conclusion, we provide evidence about how SARS-CoV-2 critically affects viral susceptibility and severity in patients with lung cancer. Our data demonstrate that lung cancer patients with up-regulated ACE2, TMPRSS2, TLR1, TLR2, and TLR6 are more likely to be susceptible to SARS-CoV-2 infection than those with down-regulated ACE2, TMPRSS2, TLR1, TLR2, and TLR6, subsequently leading to a more severe SARS-CoV-2 infection followed by promoting cancer progression through TLR2-dependent activation of NF-κB. However, it is still controversial of the role of TLR2 in lung tumor progression because TLR2 orchestrates a tumor suppressor response in early-stage lung cancer through the induction of cell-autonomous and non-cell-autonomous tumor suppressor responses. Although the precise molecular and cellular mechanisms by which TLR2 is functionally implicated in different stages of lung cancer is absolutely required, the current study gives insight into cellular and molecular mechanisms by which SARS-CoV-2 infection influences lung cancer progression in a TLR2-dependent manner. It might contribute to our understanding of the susceptibility to and the severity of SARS-CoV-2 infection in patients with lung cancer.
DECLARATIONS
AUTHOR CONTRIBUTIONS
EC and KYL conceptualized and designed the project; MJK, JYK, JHS, JS, YK, and SKJ performed experiments and analyzed data; EC, KYL, KHK, and DHK analyzed microarray data; EC and KYL wrote the manuscript; All authors read and approved the manuscript.
ACKNOWLEDGMENTS
We would like to thank Hyehwa Forum members for their helpful discussion.
FUNDING INFORMATION
This work was supported by the National Research Foundation of Korea Grants funded by the Korean Government (2023R1A2C1003762, 2021R1A2C1094478, 2021M3A912080488, and RS-2023-00217189).
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Tumor and matched normal tissues from 42 patients with primary NSCLC were obtained in accordance with the ethical principles stated in the Declaration of Helsinki. This study was approved by the Institutional Review Board of Samsung Medical Center (IRB#: 2010-07-204). We obtained written informed consent from each patient prior to surgery for using their pathological specimens for research use.
CONSENT FOR PUBLICATION
Not applicable
CONFLICT OF INTERESTS STATEMENT
The authors declare that they have no conflicts of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




