Transcription factor ETV4 promotes the development of hepatocellular carcinoma by driving hepatic TNF-α signaling
Dandan Qi and Min Lu contributed equally to this work.
Abstract
Background
Hepatic inflammation is the major risk factor of hepatocellular carcinoma (HCC). However, the underlying mechanism by which hepatic inflammation progresses to HCC is poorly understood. This study was designed to investigate the role of ETS translocation variant 4 (ETV4) in linking hepatic inflammation to HCC.
Methods
Quantitative real-time PCR and immunoblotting were used to detect the expression of ETV4 in HCC tissues and cell lines. RNA sequencing and luciferase reporter assays were performed to identify the target genes of ETV4. Hepatocyte-specific ETV4-knockout (ETV4fl/fl, alb-cre) and transgenic (ETV4Hep-TG) mice and diethylnitrosamine-carbon tetrachloride (DEN-CCL4) treatment experiments were applied to investigate the function of ETV4 in vivo. The Cancer Genome Atlas (TCGA) database mining and pathological analysis were carried out to determine the correlation of ETV4 with tumor necrosis factor-alpha (TNF-α) and mitogen-activated protein kinase 11 (MAPK11).
Results
We revealed that ETV4 was highly expressed in HCC. High levels of ETV4 predicted a poor survival rate of HCC patients. Then we identified ETV4 as a transcription activator of TNF-α and MAPK11. ETV4 was positively correlated with TNF-α and MAPK11 in HCC patients. As expected, an increase in hepatic TNF-α secretion and macrophage accumulation were observed in the livers of ETV4Hep-TG mice. The protein levels of TNF-α, MAPK11, and CD68 were significantly higher in the livers of ETV4Hep-TG mice compared with wild type mice but lower in ETV4fl/fl, alb-cre mice compared with ETV4fl/fl mice as treated with DEN-CCL4, indicating that ETV4 functioned as a driver of TNF-α/MAPK11 expression and macrophage accumulation during hepatic inflammation. Hepatocyte-specific knockout of ETV4 significantly prevented development of DEN-CCL4-induced HCC, while transgenic expression of ETV4 promoted growth of HCC.
Conclusions
ETV4 promoted hepatic inflammation and HCC by activating transcription of TNF-α and MAPK11. Both the ETV4/TNF-α and ETV4/MAPK11 axes represented two potential therapeutic targets for highly associated hepatic inflammation and HCC. ETV4+TNF-α were potential prognostic markers for HCC patients.
List of Abbreviation
-
- ETV4
-
- ETS translocation variant 4
-
- HCC
-
- hepatocellular carcinoma
-
- NASH
-
- nonalcoholic steatohepatitis
-
- HBV
-
- Hepatitis B virus
-
- Alb-Cre
-
- Cre driven by albumin promoter
-
- TNF-α
-
- tumor necrosis factor alpha
-
- IL-6
-
- interleukin-6
-
- MAPK11
-
- mitogen-activated protein kinase 11
-
- CCL4
-
- carbon tetrachloride
-
- TERT
-
- telomerase reverse transcriptase
-
- IKK-β
-
- nuclear factor kappa-B kinase subunit beta
-
- NF-κB
-
- nuclear factor kappa B
-
- STAT3
-
- signal transducer and activator of transcription 3
-
- TNFR1
-
- TNF receptor 1
-
- MYC
-
- myelocytomatosis oncogene
-
- DEN
-
- diethylnitrosamine
-
- MMP
-
- matrix metalloproteinase
-
- shRNAs
-
- short hairpin RNAs
-
- CDKN1A
-
- cyclin-dependent kinase inhibitor 1
-
- HFD
-
- high fat diet
-
- CDE
-
- choline-deficient and ethionine-supplemented diet
-
- CH
-
- cholestasis induced hepatitis
-
- YAP
-
- Yes-associated protein
-
- TEAD
-
- TEA domain family member
-
- uPA
-
- urokinase plasminogen activator
-
- CXCL1
-
- C-X-C motif chemokine ligand 1
-
- CXCL5
-
- C-X-C motif chemokine ligand 5
-
- PD-L1
-
- programmed cell death 1 ligand 1
-
- CCL2
-
- C-C motif chemokine ligand 2
-
- ROS
-
- reactive oxygen species
-
- ATCC
-
- American type culture collection
-
- GEPIA
-
- gene expression profiling interactive analysis
-
- LIHC
-
- liver hepatocellular carcinoma
-
- NCBI GEO
-
- national center for biotechnology information gene expression omnibus
-
- IHC
-
- Immunohistochemistry
-
- KEGG
-
- Kyoto encyclopedia of genes and genomes
-
- CHIP
-
- chromatin immunoprecipitation
-
- H&E
-
- hematoxylin and eosin
-
- BSA
-
- bovine serum albumin
-
- ELISA
-
- enzyme-linked immunosorbent assay
-
- ANOVA
-
- analysis of variance.
1 BACKGROUND
Liver cancer is the second most lethal cancer, with a very low 5-year survival rate. Each year, 905,000 new liver cancer cases are diagnosed, and 830,000 liver cancer-related deaths occur [1]. Liver cancer accounts for 4.7% of all new cancer cases and 8.3% of all cancer-related deaths [1]. Hepatocellular carcinoma (HCC) accounts for 80% of liver cancer cases and is the most common form of liver cancer [2, 3]. Hepatitis B virus (HBV) and hepatitis C virus infection, alcohol abuse, obesity and diabetes-associated nonalcoholic steatohepatitis (NASH) are the main risk factors for HCC [4, 5]. Although the incidence of alcoholic hepatitis and metabolic syndrome-related NASH is increasing faster in western countries, HBV infection remains the major risk factor for HCC in China [6, 7].
Sustained inflammation accelerated the development and aggressiveness of HCC by promoting the survival of hepatocytes [8, 9], enhancing angiogenesis [10], increasing HCC escape of immune surveillance [11], and enhancing invasion and metastasis [12, 13]. Interleukin-6 (IL-6) produced by Kupffer cells activated the signal transducer and activator of transcription 3 (STAT3) pathway in hepatocytes and induced the transcription of myelocytomatosis oncogene (MYC), B-cell lymphoma-2 (BCL-2) and Cyclin B to promote the proliferation and survival of hepatocytes [14, 15]. Tumor necrosis factor-alpha (TNF-α) activated the downstream inhibitor of nuclear factor kappa-B kinase subunit beta (IKK-β)/nuclear factor kappa B (NF-κB) signaling pathway and increased the number and size of diethylnitrosamine (DEN)-induced mouse HCC tumors [16]. Lymphotoxin α/β expression induced chronic liver injury and hepatitis, which further stimulated the compensatory proliferation of hepatocytes and led to tumorigenesis [17]. Both dietary and genetic obesity enhanced liver inflammation and carcinogenesis by inducing TNF-α and IL-6 expression [14, 15]. TNF receptor 1 (TNFR1)-deficient mice had decreased tumor formation than wild-type mice when DEN and a high-fat diet (HFD) were used to induce HCC tumorigenesis [18]. Inflammation played a key role in HCC development, and inflammatory cytokines were reported mainly expressed in immune cells. It has been reported that inflammatory cell-secreted TNF-α played a critical role in inducing chronic inflammation of the liver [19-21], and macrophage infiltration had important impact on tumor progression [22]. However, the mechanisms of hepatocytes secreting TNF-α during HCC development and efficacy of TNF-α on regulating macrophages infiltration were unclear.
ETV4 is a transcription factor which belongs to the E26 transformation-specific (ETS) family. All the members of this family have a conserved ETS DNA-binding motif. It has been reported that ETV4 played an important role in tissue development, such as neuronal differentiation and kidney morphogenesis [23]. ETV4 was also defined as an oncogene that promoted the growth and metastasis of gastric cancer and the development of prostate cancer [24]. However, the detailed mechanism of how ETV4 regulating inflammation is not fully elucidated.
In this study, we focused on the function of ETV4 in liver inflammation and HCC development. We investigated the association of ETV4 with HCC formation by analyzing the clinical samples and diethylnitrosamine (DEN)-carbon tetrachloride (CCL4)-induced HCC mice. We explored the functions of ETV4 in liver inflammation via acute and chronic liver inflammation mouse model. These studies were expected to uncover the mechanisms of ETV4 regulating liver inflammation and HCC formation.
2 MATERIALS AND METHODS
2.1 Cell lines
The human embryonic kidney cell line 293T were purchased from American Type Culture Collection (ATCC) (Manassas, VA, USA). The human hepatoma cell line HepG2 were purchased National Collection of Authenticated Cell Cultures (Shanghai, China). The HBV transgenic cell line HepG2-4D14 and human hepatoma cell line Huh7 were gifts from Dr. Dongping Xu (The Fifth Medical Center of Chinese PLA General Hospital, Beijing, China). The Huh7 cell was verified by short tandem repeat sequence in 2023. All the cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM, 12800-017, Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS, ST30-3302, PAN-Biotech, Aidenbach, Germany).
2.2 Datasets and analysis
The differentially expressed genes (DEGs) in HCC tissues compared to normal tissues were downloaded from Gene Expression Profiling Interactive Analysis (GEPIA) in which the RNA expression data from the Cancer Genome Atlas (TCGA, http://gepia.cancer-pku.cn/) were analyzed, and they were overlapped with the DEGs in HepG2-4D14 compared to HepG2 cells. The Cancer Genome Atlas Liver Hepatocellular Carcinoma (TCGA-LIHC) datasets were downloaded from TCGA, and the ETV4 mRNA level in tumors and normal tissues was analyzed. GSE64041, GSE83148, GSE94660 and GSE84005 were obtained from National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) profiles, and the relative ETV4 mRNA level in these cohorts was analyzed. JASPAR (https://jaspar.genereg.net/) was used to predict the ETV4-binding sites on the promoter of TNF-α or MAPK11. HCC single-cell transcriptome data were from public database (https://db.cngb.org/PRHCCdb/), in which the expression profiles of tumor cells and different kinds of immune cells were obtained [25]. xCell (https://comphealth.ucsf.edu/app/xcell) was used to perform cell type enrichment analysis from gene expression data, and enumerate immune cell subsets from TCGA-LIHC transcriptomes [26].
2.3 Patient samples
Twenty-seven HCC tumor tissues and paired peritumoral tissues were defined as Cohort 1 in which the patients were underwent surgery between August 2012 and December 2014. Total RNA was extracted from all the tumor tissues and nontumor tissues and analyzed by quantitative real-time PCR (qRT-PCR) to measure ETV4 mRNA levels. A total of 128 formalin-fixed paraffin-embedded (FFPE) primary HCC liver specimens were defined as Cohort 2 and subjected to Immunohistochemistry (IHC) staining for ETV4, TNF-α, MAPK11 and CD68. Patients in Cohort 2 were hospitalized and underwent surgery between Jun 2012 and Dec 2013. The patients were all diagnosed with primary HCC, with no cholangiocarcinoma or other cancers. Patients in Cohort 2 were divided into HBV-low (≤40 DNA copies/mL) and HBV-high (>40 DNA copies/mL) groups according to the serum HBV DNA copy number before surgery. Sixty-six HCC patients who underwent surgery between July 2012 and November 2013 were defined as Cohort 3. The patients in Cohort 3 were divided into HBV-negative (both HBsAg and HBeAg were negative, and HBV DNA was undetectable in serum) and HBV-positive groups. The clinical characteristics of the enrolled patients are listed in Supplementary Table S1. All the patient samples were collected from the Fifth Medical Center of Chinese PLA General Hospital. All the participants provided written informed consent. The patient samples were assigned arbitrary identification numbers based on the order of enrollment.
For the single-marker overall survival (OS) analyses, the quartile was used to set the group cutoff according to the levels of ETV4, TNF-α or MAPK11. The top 25% was classified as high, the bottom 25% was classified as low, and the medium 50% was classified as intermediate.
For the double-marker OS analyses, firstly the medium was used to set the group cutoff according to the levels of ETV4, TNF-α or MAPK11, in which the top 50% was classified as high, the other 50% was classified as low. Then, patients with both ETV4-high and TNF-α-high were defined as ETV4+TNF-α-high, while patients with both ETV4-low and TNF-α-low were defined as ETV4+TNF-α-low. The other patients were classified as intermediate. The same group classification was applied for ETV4+MAPK11 groups.
2.4 Real-time quantitative PCR (qRT-PCR)
The human and mouse liver tissues were homogenized with the tissue grinder. Total RNA was extracted from tissues and cell lines with TRIzol (1559608, Invitrogen, Carlsbad, CA, USA). cDNA was synthesized with Hifair II 1st strand cDNA Synthesis SuperMix (11123ES60, YEASEN, Shanghai, China). The random and oligo(dT)s primer mixtures were used for the cDNA synthesis. The reaction mixture was pre-denatured for 5 min at 95°C, then denatured for 10 s at 95°C, annealed and extended for 30 s at 60°C, for a total 40 cycles. qRT-PCR was performed with qPCR SYBR Green Master Mix (11202ES08, YEASEN, Shanghai, China). The primers specific for the indicated genes used in qRT-PCR are listed in Supplementary Table S2.
2.5 RNA sequencing and analysis
The human ETV4 gene was cloned into the lentiviral vector pLL-IRES-puro (provided by Dr. Wenjun Liu, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China) and named as pLL-ETV4. Short hairpin RNAs (shRNAs) targeting human ETV4 were cloned into the pSIH1-GFP lentiviral plasmid. The pLL-ETV4 vector or the pSIH1-GFP-shETV4 vector and the lentivirus packing plasmids were co-transfected into 293T cells. Lentivirus was harvested 72 h post transfection. HepG2 cells were infected with lentivirus carrying ETV4 to build the ETV4-overexpressing HepG2 cells (HepG2-ETV4-oe). Huh7 cells were infected with lentivirus carrying ETV4 shRNA to build the ETV4-knockdown Huh7 cells (Huh7-shETV4). Total RNA was extracted from HepG2-ETV4-oe and control cells or Huh7-shETV4 and control cells. The mRNAs were enriched and subjected to mRNA library construction and sequencing by BGI (BGISEQ-500 platform, Shenzhen, China). The raw reads were filtered and quantity controlled to obtain clean reads. Then, the clean reads were aligned with the Homo sapiens genome (GCF_000001405.37_GRCh38.p11). The gene abundance was measured in fragments per kilobase of exon per million fragments mapped (FPKM). The genes with fold change higher or lower than 2 in HepG2-ETV4-oe vs HepG2 cells, or Huh7-shETV4 vs. Huh7 cells, were defined as DEGs. The DEGs within the groups were further analyzed with Kyoto Encyclopedia of Genes and Genomes (KEGG) for signaling pathway enrichment.
2.6 Dual-luciferase reporter assay
Human TNF-α and MAPK11 promoters and their mutants were cloned into the pGL4 basic vector. HEK293T cells were seeded in 24-well plates (3×105 cells per well) and transfected with the pLL-ETV4 or control vector together with the pGL4-TNF-α/MAPK11 promoter and pRL-TK Renilla reporter plasmids as an internal control for 24 h. Then, the cell lysates were harvested and subjected to luciferase assay with the dual-specific luciferase reporter assay system (E1980, Promega, Madison, WI, USA).
2.7 Chromatin immunoprecipitation (CHIP) assay
The CHIP assay was performed with the CHIP assay kit (P2078, Beyotime, Wuhan, Hubei, China). Briefly, Huh7 cells were cross-linked with 4% formaldehyde for 10 min, then treated with 125 mmol/L glycine for 5 min. Cells were collected and sonicated. The samples were centrifuged for 10 min at 8,000 g at 4°C. The supernatants were subjected to immunoprecipitation (IP) with IgG or anti-ETV4 antibodies for 4 h, followed by incubation with protein A Sepharose beads for 2 h. The IP samples were eluted and treated with proteinase K, and then the DNA were extracted with phenol-chloroform and subjected to qRT-PCR with indicated primers.
2.8 Mouse models
C57BL/6 ETV4 conditional knockout mice (ETV4fl/fl, alb-cre) were generated with the CRISPR/Cas9-guided homologous recombination system (Biocytogen, Beijing, China). Briefly, an ETV4 targeting vector containing the ETV4 genome sequence from exon 6 to exon 13 with 5’loxp and 3’loxp flanking sites was constructed. Small guide RNA (sgRNA) sequences that effectively targeted intron 6 and the downstream sequence of exon 13 were selected with the UCATM assay, and Cas9/sgRNA plasmids were constructed. The ETV4 targeting vector and Cas9/sgRNA plasmids were microinjected into zygotes from the oviducts of 4-week-old female C57BL/6 mice. The genotype of 7-day-old male and female ETV4fl/fl mice was confirmed by Southern blotting and DNA sequencing. For Southern blotting, the genomic DNA was digested with AseI, and then subjected to hybridization with digoxin-labeled probe of ETV4 exon 5. Hepatocyte-specific ETV4fl/fl, alb-cre mice were generated by crossing ETV4fl/fl mice with Alb-Cre mice.
To generate hepatocyte-specific ETV4 conditional transgenic mice (ETV4Rosa26-floxp-gfp-floxp-mETV4/+, alb-cre, ETV4Hep-TG), floxp-gfp-polyA-floxp mouse ETV4 (mETV4) was inserted into the Rosa26 locus of C57BL/6 wild type mice. Rosa26 is a non-coding gene on chromosome 6 of the mice. In general, the gene inserted into Rosa26 locus can be expressed normally while did not affect the expression of neighboring genes. The ETV4Hep-TG mice were generated by crossing ETV4Rosa26-floxp-gfp-floxp-mETV4/+ mice with Alb-Cre mice.
HCC mouse model was generated as follows: 2-week-old male ETV4fl/fl and ETV4fl/fl, alb-cre or wild type and ETV4Hep-TG mice were intraperitoneally injected with 25 mg/kg DEN (N0756, Sigma Aldrich, Saint Louis, MO, USA), followed by intraperitoneally injection of 0.5 mL/kg CCL4 (C0731530924, Nanjing Reagent, Nanjing, China) every two weeks. The mice were euthanized via cervical dislocation at the 21-week-old. The livers were harvested and imaged, and the tumor number and size were measured. Hematoxylin and eosin (H&E) staining was performed and scanned by Leica digital pathology scanner Aperio CS2 (Wetzlar, Germany). The numbers of microscopic tumors were counted. The diameter of the tumor and the total tissue area were measured by Image Scope. Numbers of tumors per cm2 were calculated.
For the acute inflammation model, 8-week-old male wild type and ETV4Hep-TG mice were intraperitoneally injected with DEN (100 mg/kg) for 8 h and euthanized. Then the livers were fixed with formalin and subjected to IHC staining.
To establish chronic inflammation model, 2-week-old male ETV4fl/fl and ETV4fl/fl, alb-cre or wild type and ETV4Hep-TG mice were intraperitoneally injected with a single dose of DEN (25 mg/kg), followed by CCL4 (0.5 mL/kg) treatment every two weeks, and euthanized at 16 weeks. The livers were fixed with formalin and subjected to H&E staining or IHC staining with the indicated antibodies.
To deplete macrophages, 8-week-old male wild type and ETV4Hep-TG mice were intravenously injected with 10 mg/kg GdCl3 (203289, Sigma Aldrich, Saint Louis, MO, USA) every two days for 3 times [27, 28].
All the mice in this study were housed under specific pathogen-free (SPF) conditions at 25°C with 50% humidity. The mice were euthanized via cervical dislocation at the end of experiments. All the experiments were approved by the Ethical Review Committee of the Institute of Microbiology, Chinese Academy of Sciences (APIMCAS2019009).
2.9 IHC analysis
The human and mouse FFPE samples were sliced in 5 mm thickness and subjected to standard IHC procedures. In brief, the sections were treated with xylene firstly, and then hydrated in series of ethanol (99.9% ethanol, 95% ethanol, 75% ethanol, 50% ethanol, H2O). The antigen retrieval was performed as follows: for ETV4 and TNF-α, the sections were treated with 1 mmol/L EDTA/10 mmol/L Tris HCl (pH 9.0); for MAPK11 and CD68, the sections were treated with 10 mmol/L sodium citrate (pH 6.0) for 30 min. The sections were blocked with 5% bovine serum albumin (BSA) (0032, Amresco, Solon, OH, USA) and incubated with primary antibodies (anti-ETV4, 1:50, overnight at 4°C; anti-MAPK11, 1:125, 2.5 h at room temperature; anti-TNF-α, 1:500, 2.5 h at room temperature; anti-CD68, 1:300, overnight at 4°C) and then incubated with horseradish peroxidase (HRP)-conjugated secondary antibody (PV-9000, ZSGB-BIO, Beijing, China) for 40 min at room temperature. The whole slide image was scanned by a NanoZoomer 2.0 HT (Hamamatsu, Japan) and analyzed with a HALO 2.3 digital pathology analysis system (Indica Labs, NM, USA). The tumor and adjacent nontumor areas were manually defined by using the HALO software (Albuquerque, NM, USA). The average density and percentage of positive cells for H-score was calculated with the equation: H-score = weak positive cell percentage × 1 + moderately positive cell percentage × 2 + strongly positive cell percentage × 3.
2.10 Primary mouse hepatocyte isolation
Mouse primary hepatocytes were isolated according to the procedures of Wen Zhang's Lab (http://www.mouselivercells.com/). Briefly, 8-week-old wild type or ETV4Hep-TG male mice were anesthetized by intraperitoneally injection of 400 μg/g 2,2,2-tribromoethanol (T48402, Sigma Aldrich, Saint Louis, MO, USA), and then perfused with 100 units/mL type IV collagenase (17104, Gibco, Carlsbad, CA, USA) via the portal vein for 3-5 min until the livers were digested sufficiently. The livers were dissected out and teared apart to release the hepatocytes. The hepatocytes were collected after centrifugation (3 min, 50 g at 4°C) and plated. The ETV4, TNF-a and MAPK11 mRNA levels were measured by qRT-PCR.
2.11 Enzyme-linked immunosorbent assay (ELISA)
HepG2 cells overexpressing ETV4 or control cells were seeded in 24-well plates (2×105 cells per well) for 36 h. Then, the supernatants were collected and subjected to ELISA with a human TNF-α ELISA kit (KIT10602, Sino Biological, Beijing, China).
2.12 Flow cytometry analysis
The mouse livers were minced into 1 mm3 pieces, followed by digestion with collagenase for 30 min. The cells were centrifuged at 350 g at 4°C for 3 min, and purified in 40% percoll. Subsequently, the cells were stained with anti-CD68 (137009, FA-11), anti-F4/80 (123110, BM8), anti-CD45 (103115, 30-F11), anti-CD11b (101212, M1/70), anti-CD3 (100205, 17A2), anti-CD4 (100515, RM4-5), anti-CD8 (100733, 53-6.7), anti-Ly6G (127617, 1A8) antibodies (BioLegend, San Diego, CA, USA), and sorted using a BD FACSAria II Cell Sorter (BD Bioscience, San Jose, CA, USA).
2.13 Statistical analysis
The OS of HCC patients after diagnosis was analyzed with the Kaplan-Meier estimator and calculated with the log-rank test. Student t test was used for two-group comparisons, and two-way analysis of variance (ANOVA) was used for multiple-group comparisons. P < 0.05 was considered significance.
3 RESULTS
3.1 The level of ETV4 was negatively associated with the survival of HCC patients
To investigate the HBV-related genes involved in the development of HCC, we overlapped the DEGs in the HBV transgenic cell line HepG2-4D14 compared to the parental cell line HepG2 and the DEGs in HCC tumor tissues compared to normal tissues in the TCGA-LIHC cohort (Figure 1A). Sixty-nine overlapped genes were identified (Supplementary Table S3), and the differential expression pattern is shown in Figure 1B. Among these 69 genes, ETV4 expression level was the most significantly associated with the OS of HCC patients (Supplementary Figure S1A), and the mRNA level of ETV4 was much higher in tumor tissues than in normal tissues according to the data from TCGA (Supplementary Figure S1B) and GEO HCC datasets (Supplementary Figure S1C). We next compared the mRNA level of ETV4 in 27 pairs of HCC tissues and their paired adjacent peritumoral tissues, and found that ETV4 was upregulated in HCC tumor tissues comparing to their paired adjacent peritumoral tissues (Figure 1C). The HCC datasets from GEO also showed that ETV4 was higher in tumors than in their paired peritumoral tissues (Supplementary Figure S1C-E). Then, we performed IHC staining of ETV4 in 128 HCC liver tissues which were collected from patients who underwent surgery between 2012 and 2013. The mean intensity of ETV4 staining was evaluated and representative images are shown in Figure 1D. Kaplan-Meier analysis demonstrated that patients with higher ETV4 levels had poorer survival rate (Figure 1E), which was consistent with the results shown in Supplementary Figure S1A, and both the percentage of ETV4-positive cells (Figure 1F) and the H-score (Supplementary Figure S1F) were higher in tumor tissues than in paired adjacent peritumoral tissues. All these data demonstrated that ETV4 was highly expressed in HCC tumor tissue, indicating that ETV4 level might predict poor survival rate of HCC patients.
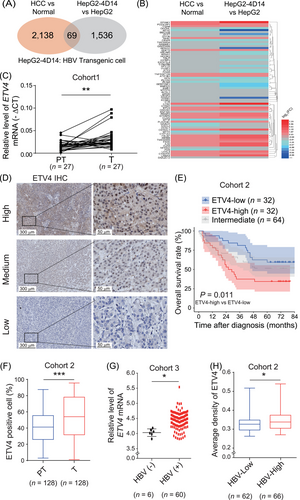
ETV4 is highly expressed in HCC and HBV-positive liver tissues, and its expression is negatively correlated with the survival of HCC patients. (A) Total RNA was extracted from HepG2 and HBV transgenic cell lines HepG2-4D14 and subjected to RNA deep sequencing analysis. The differentially expressed genes in HepG2-4D14 cells compared to HepG2 cells overlapped with the abnormally expressed genes in HCC tumors compared to normal tissues in the TCGA-LIHC cohort. (B) Heatmap of the overlapping genes in A. (C) The ETV4 level in 27 pairs of HCC tissues and their adjacent nontumoral tissues was determined by qRT-PCR. (D-F) A total of 128 paraffin-embedded primary HCC specimens were subjected to IHC staining with an anti-ETV4 antibody. The intensity of ETV4 staining was quantitatively scored with the HALO digital pathology system, and samples were classified into three subgroups (high, medium and low) according to their scores. Quartile was used to set the group cutoff. Representative images are shown in D. The overall survival of patients analyzed with Kaplan-Meier analysis is shown in E. The medium was classified as intermediate group. The percentage of ETV4-positive cells in peritumoral and tumor regions in each sample according to IHC staining was calculated by the HALO digital pathology system (F). (G) The relative mRNA level of ETV4 in HBV-negative (n = 6) and HBV-positive (n = 60) HCC tissues was measured by qRT-PCR. (H) HCC patients were divided into two groups (HBV-low, n = 62; HBV-high, n = 66) according to the HBV viral load in their serum. The ETV4 protein levels in HCC tissues were measured by IHC, and the average intensity of ETV4 staining was quantified using the HALO digital pathology system. In the Box and whiskers plots, boxes represented 25-75% percentile, whiskers were min to max, and division line was median. In the scatter plots, division line was mean, whiskers were Standard deviation. * P < 0.05, ** P < 0.01, *** P < 0.001.
Abbreviations: ETV4, ETS translocation variant 4; HBV, hepatitis B virus; HepG2-4D4, HBV Transgenic cell; HCC, hepatocellular carcinoma; TCGA, The Cancer Genome Atlas; LIHC, liver hepatocellular carcinoma; qRT-PCR, real-time quantitative PCR; PT, peritumoral tissue; T, tumor; IHC, immunohistochemistry.
3.2 ETV4 expression was upregulated in HBV-positive HCC and chronic hepatitis liver tissues
Both ETV4 mRNA and protein levels were higher in HepG2-4D14 cells than in parental HepG2 cells (Supplementary Figure S1G-H), which is consistent with the results in Figure 1B. We analyzed the relative ETV4 level in the GEO dataset (GSE83148), which included gene expression data from 122 liver samples from chronic hepatitis B (CHB) patients and 6 liver samples from healthy controls. The results indicated that the mRNA level of ETV4 was higher in the CHB samples than in the normal samples (Supplementary Figure S1I). Also, the mRNA levels of ETV4 were higher in tumor tissues from HBV-positive patients than in those from HBV-negative patients in Cohort 3 (Figure 1G). We then divided the HCC samples in Cohort 2 into two groups (HBV-low and HBV-high) according to the HBV DNA copy numbers in the serum of patients before surgery. The data of IHC staining demonstrated that the protein level of ETV4 was higher in tumor tissues from patients with a higher HBV load than in those from patients with a lower HBV load (Figure 1H). These data suggested that the ETV4 level was upregulated in HBV-related HCC.
In addition, we analyzed the TCGA dataset and found that the level of ETV4 mRNA was also higher in alcohol or non-alcoholic fatty liver disease-associated HCC tumors than in normal tissues (Supplementary Figure S1J).
3.3 ETV4 directly promoted the transcription of TNF-α and MAPK11
To identify downstream genes regulated by ETV4, we compared the transcription profiles of ETV4-overexpressing HepG2 cells (HepG2-ETV4-oe) and control cells (HepG2-Ctrl) and ETV4-knockdown Huh7 (Huh7-shETV4) cells and control cells (Huh7-shR) by RNA sequencing. We overlapped the DEGs in the 2 sets described above and identified 159 potential ETV4 downstream genes (Figure 2A-B). By using KEGG pathway analysis, we found that ETV4-upregulated genes were mainly involved in TNF, NF-κB and MAPK signaling pathways (Figure 2C). TNF-α is an inflammatory cytokine that abnormally expressed in many kinds of chronic or acute liver injury/inflammation [17, 29, 30]. MAPK11 (p38β) belongs to the family of the p38 MAPKs, which can induce the production of TNF-α and IL-1β [31-33]. Therefore, TNF-α and MAPK11 were evaluated in priority as they played critical roles in inflammation.
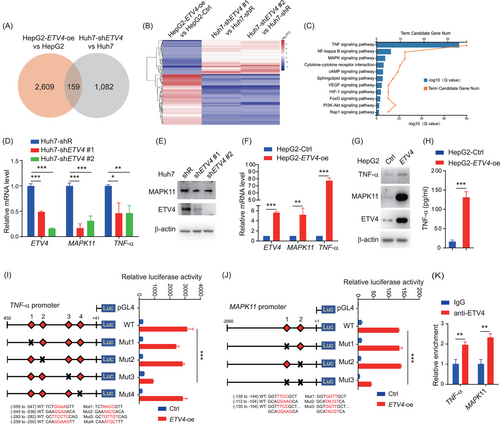
ETV4 regulates the transcription of TNF-α and MAPK11. (A, B) HepG2-Ctrl and ETV4-overexpressing HepG2 (HepG2-ETV4-oe), Huh7-shR and ETV4 knockdown Huh7 (shETV4#1, shETV4#2) cells were harvested, and total RNA was extracted and subjected to RNA sequencing. The differentially expressed genes between HepG2 and HepG2-ETV4-oe cells or between Huh7-shR and Huh7-shETV4 cells are shown in a Venn diagram (A) and heatmap (B). (C) KEGG pathway analysis was performed with the genes whose expression levels were upregulated in HepG2-ETV4-oe cells compared to HepG2 cells. (D, E) Total RNA extracted from Huh7-shR, Huh7-shETV4#1, and Huh7-shETV4#2 cells were subjected to qRT-PCR to quantify the mRNA levels of ETV4, MPAK11 and TNF-α, three replicates were performed (D). The protein levels of ETV4 and MAPK11 in the cells mentioned above were determined by immunoblotting with actin as the internal control, two replicates were performed (E). (F-H) Total RNA extracted from HepG2-Ctrl and HepG2-ETV4-oe cells was subjected to qRT-PCR to measure the mRNA levels of ETV4, MAPK11 and TNF-α, three replicates were performed (F). TNF-α and MAPK11 protein levels in the cells mentioned above were measured by immunoblotting with the indicated antibodies, three replicates were performed (G). TNF-α levels in the supernatant were measured by ELISA, four replicates were performed (H). (I) 293T cells were transfected with pLL-ETV4 and the TNF-α promoter luciferase reporter (WT) or a luciferase reporter with the potential ETV4 binding sites mutated (Mut1-4) for 24 h. The cell lysates were harvested and subjected to luciferase assay. Two replicates were performed. (J) 293T cells were transfected with pLL-ETV4 and the MAPK11 promoter luciferase reporter (WT) or a luciferase reporter with the potential ETV4 binding sites mutated (Mut1-3) for 24 h. The cell lysates were harvested and subjected to luciferase assay. Three replicates were performed. (K) CHIP assay was performed in Huh7 cells with anti-ETV4 antibodies or anti-IgG control. qRT-PCR was used to quantify the relative fold change of ETV4 on TNF-α or MAPK11 promoter. Two replicates were performed. The column bar graphs were presented as the mean ± SD. * P < 0.05, ** P < 0.01, *** P < 0.001.
Abbreviations: ETV4, ETS translocation variant 4; TNF-α, tumor necrosis factor alpha; MAPK11, mitogen-activated protein kinase 11; oe, overexpression; shR, control shRNA; Ctrl, control; WT, wild type; Mut, mutation; ELISA, enzyme-linked immunosorbnent assay; CHIP, chromatin immunoprecipitation; qRT-PCR, real-time quantitative PCR; Luc, luciferase.
First, we confirmed that ETV4 knockdown in Huh7 cells caused a reduction in the mRNA levels of TNF-α and MAPK11 (Figure 2D) as well as the protein level of MAPK11 (Figure 2E). In contrast, overexpression of ETV4 in HepG2 cells enhanced both the mRNA and protein levels of TNF-α and MAPK11 (Figure 2F-H). We predicted the ETV4-binding sites on the promoters of TNF-α and MAPK11 on the JASPAR website. Then, we generated the luciferase reporters for the promoters of TNF-α and MAPK11 and performed luciferase assays. The data showed that ETV4 could significantly enhance the activities of the TNF-α and MAPK11 promoters (Figure 2I-J). Furthermore, we also performed luciferase assays with the promoters of TNF-α or MAPK11 in which the potential ETV4-binding sites were mutated, and identified the regions in the TNF-α and MAPK11 promoters that were critical for ETV4 binding (Figure 2I-J). In addition, we performed the CHIP assay, and the data showed that ETV4 could bind to the promoters of the TNF-α and MAPK11 directly in Huh7 cells (Figure 2K). In conclusion, ETV4 can directly promote the transcription of TNF-α and MAPK11.
3.4 ETV4 enabled macrophage accumulation and facilitated liver inflammation
To test whether ETV4 could regulate the expression of TNF-α and MAPK11 and enhance the liver inflammation in vivo, we generated hepatocyte-specific ETV4-transgenic mice (named as ETV4Hep-TG) by crossing ETV4Rosa26-floxp-gfp-floxp-mETV4/+ mice with Alb-Cre mice (Figure 3A and Supplementary Figure S2A). The genotype of ETV4Hep-TG was confirmed by PCR (Supplementary Figure S2B) and ETV4 protein was determined by immunoblotting (Figure 3B). We also generated hepatocyte-specific ETV4-knockout mice (ETV4fl/fl, alb-cre) by crossing ETV4fl/fl with Alb-Cre mice (Figure 3C-D and Supplementary Figure S2C-D). No significant changes of liver/body ratio were observed in 8-week-old ETV4 transgenic or ETV4 knockout mice (Supplementary Figure S2E-F).
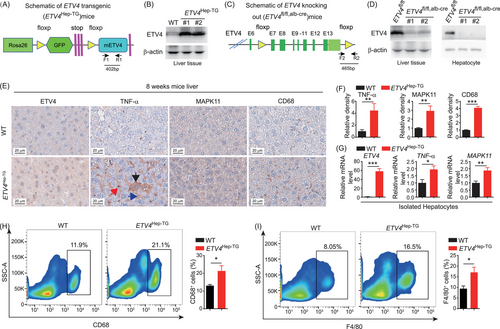
ETV4 upregulates TNF-α/MAPK11 expression and liver inflammation. (A) The genomic map of conditional ETV4-transgenic mice (ETV4Rosa26-floxp-gfp-floxp-mETV4/+). The F1 and R1 primers were used for genotyping. (B) ETV4Rosa26-floxp-gfp-floxp-mETV4/+ mice were crossed with hepatocyte-specific Cre mice to generate ETV4Hep-TG (ETV4Rosa26-floxp-gfp-floxp-mETV4/+, alb-cre) mice. The cell lysates from liver tissues of 8-week-old WT or ETV4Hep-TG mice were subjected to immunoblotting with an anti-ETV4 antibody. (C) The genomic map of conditional ETV4-knockout mice (ETV4fl/fl, alb-cre); F2 and R2 represent the primers used for genotyping. (D) The lysates from the total liver tissues (left) and hepatocyte isolated by perfusion (right) of 8-week-old ETV4fl/fl or ETV4fl/fl, alb-cre mice were subjected to immunoblotting with an anti-ETV4 antibody. (E, F) WT (n = 5) or ETV4Hep-TG (n = 3) mice were sacrificed at 8 weeks of age, and liver tissues were harvested and subjected to IHC staining of TNF-α, MAPK11 and CD68. Representative images are shown (E). Black arrow, TNF-α positive hepatocyte; Blue arrow, TNF-α in intracellular area; Red arrow, TNF-α positive immune cell. The intensity of TNF-α, MAPK11 and CD68 staining was quantified (F). (G) Hepatocytes were isolated from WT (n = 7) or ETV4Hep-TG (n = 8) mice. The relative mRNA level of ETV4, TNF-α and MAPK11 were determined by qRT-PCR. (H, I) The proportions of CD68+ (H) and F4/80+ (I) cells were analyzed by flow cytometry in 8-week-old WT and ETV4Hep-TG mice liver. The column bar graphs were presented as the mean ± SD. * P < 0.05, ** P < 0.01, *** P < 0.001.
Abbreviations: ETV4, ETS translocation variant 4; TNF-α, tumor necrosis factor alpha; MAPK11, mitogen-activated protein kinase 11; WT, wild type; Alb-Cre, cre driven by albumin promoter; IHC, immunohistochemistry; qRT-PCR, real-time quantitative PCR.
The protein levels of hepatic TNF-α, MAPK11 and CD68 were examined. The data showed that levels of TNF-α, MAPK11 and CD68 were higher in ETV4Hep-TG than that in wild type (WT) mice (Figure 3E-F). To further explore whether ETV4 enhanced the transcription of TNF-α and MAPK11 in hepatocytes, we isolated primary hepatocytes from the WT or ETV4Hep-TG mice, and found that the mRNA levels of TNF-α and MAPK11 in hepatocytes were significantly higher in ETV4Hep-TG than that in WT mice (Figure 3G). We further performed the flow cytometry analysis and found that both CD68+ and F4/80+ cells were significantly enriched in ETV4Hep-TG than in WT mice (Figure 3H-I). The neutrophils were also higher in ETV4Hep-TG mice than in WT mice, but there was no difference of CD4+ T cells and CD8+ T cells between WT and ETV4Hep-TG mice (Supplementary Figure S3). Taken together, these results indicated that ETV4 promoted the expression of TNF-α and MAPK11 in hepatocytes and the accumulation of macrophages and neutrophils in the liver.
To investigate whether ETV4 promoted the expression of TNF-α and MAPK11 and the accumulation of macrophages during liver inflammation, we measured the protein levels of TNF-α, MAPK11 and CD68 in the livers of control mice and mice treated with DEN to induce acute inflammation and found that TNF-α, MAPK11 and CD68 were expressed higher in the ETV4Hep-TG mice than in the WT mice (Figure 4A-B). We also performed macrophage depletion with GdCl3 in mice treated with DEN and found that TNF-α expression was still higher in ETV4Hep-TG than in WT mice (Supplementary Figure S4A-B). The above data indicated that ETV4 could upregulate TNF-α expression and enhance macrophage accumulation in the liver during acute inflammation. To uncover the effect of ETV4 during chronic liver injury and inflammation, we treated the mice with DEN combined with CCL4 (Figure 4C). IHC staining showed that the levels of TNF-α, MAPK11 and CD68 in the livers of the ETV4Hep-TG mice were higher than those in WT mice (Figure 4D-E), indicating that ETV4 could trigger liver inflammation and macrophage accumulation via upregulating TNF-α and MAPK11 expression when there was chronic liver injury. Otherwise, knockout of ETV4 caused a marked reduction in the TNF-α, MAPK11 and CD68 levels (Figure 4F-G). To completely exclude the possible influence of acute liver injury caused by CCl4, we further measured the protein levels of TNF-α, MAPK11 and CD68 in the liver samples from the mice 7 days after the final injection of CCL4 (Supplementary Figure S4C) and obtained consistent results in the ETV4Hep-TG mice (Supplementary Figure S4D-E) and the ETV4fl/fl, alb-cre mice (Supplementary Figure S4F-G). Overall, these results demonstrated that ETV4 enabled macrophage accumulation and facilitated liver inflammation via modulating the ETV4-TNF-α and ETV4-MAPK11 axis.
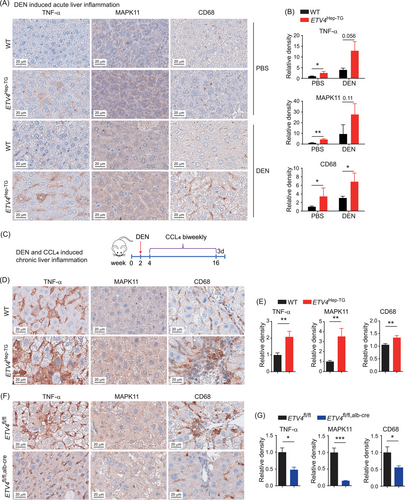
ETV4 enhances macrophage accumulation and inflammation in liver under injury. (A, B) Eight-week-old WT or ETV4Hep-TG mice were intraperitoneally injected with DEN (100 mg/kg) or PBS for 8 h, and the livers were harvested and subjected to IHC staining to measure the protein levels of TNF-α, MAPK11 and CD68 (A). The intensities of the proteins mentioned above were calculated (B). WT PBS (n = 4), ETV4Hep-TG PBS (n = 4), WT DEN (n = 4), ETV4Hep-TG DEN (n = 4). (C) Schematic of DEN-CCL4 induced chronic liver inflammation in mice. (D-G) Wild-type and ETV4Hep-TG or ETV4fl/fl and ETV4fl/fl, alb-cre mice were treated as shown in C. The liver tissues were harvested from the sacrificed mice and subjected to IHC staining for TNF-α, MAPK11 and CD68. Representative images are shown in D, F. The intensities of TNF-α, MAPK11 and CD68 staining were quantified (E, G). WT (n = 8), ETV4Hep-TG (n = 8), ETV4fl/fl (n = 5), ETV4fl/fl, alb-cre (n = 7). The column bar graphs were presented as the mean ± SD. * P < 0.05, ** P < 0.01, *** P < 0.001.
Abbreviations: ETV4, ETS translocation variant 4; TNF-α, tumor necrosis factor alpha; MAPK11, mitogen-activated protein kinase 11; DEN, diethylnitrosamine; PBS, phosphate buffered saline; WT, wild type; Alb-Cre, cre driven by albumin promoter; CCL4, carbon tetrachloride; IHC, immunohistochemistry.
The data showed that ETV4 could promote the inflammation in the liver. In order to know whether inflammation could influence the level of ETV4, we treated the mice with DEN or CCL4, and observed that the mRNA level of ETV4 in the liver was increased after treatment with DEN or CCL4 (Supplementary Figure S5), indicating that inflammation may also trigger the transcription of ETV4.
3.5 ETV4 promoted liver tumorigenesis
Chronic inflammation plays an important role in HCC development. To identify whether ETV4 could promote liver tumorigenesis in vivo, we established the DEN-CCL4-treated HCC model in transgenic mice (Figure 5A). After DEN-CCL4 treatment, the number of total surface tumors and the number of tumors larger than 1 mm3 were lower in the livers of the ETV4fl/fl, alb-cre mice than in those of the ETV4fl/fl mice (Figure 5B-D), and the liver/body weight ratio was also lower in the ETV4fl/fl, alb-cre mice than in the ETV4fl/fl mice (Figure 5E). In addition, the number of microscopic tumors in the ETV4fl/fl, alb-cre mice were lower than that in the ETV4fl/fl mice (Figure 5F-H). These data indicated that knockout of ETV4 in hepatocytes suppressed liver tumor formation in DEN-CCL4 HCC model.
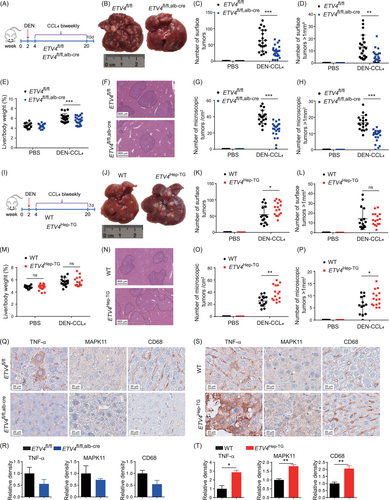
ETV4 promotes DEN-CCL4-induced HCC formation. (A) Schematic of DEN-CCL4-induced HCC in ETV4fl/fl and ETV4fl/fl, alb-cre mice. (B-H) The liver tissues from sacrificed mice as shown in A were harvested. Representative images of the livers of ETV4fl/fl and ETV4fl/fl, alb-cre mice are shown (B). The total number of surface tumors and the number of surface tumors larger than 1 mm3 were calculated (C, D). The liver/body weight of the mice was measured (E). The livers were fixed and subjected to H&E staining. Representative images are shown (F). The total number of microscopic tumors per cm2 and the number of microscopic tumors larger than 1 mm2 per cm2 were calculated according to the H&E staining in F (G, H). ETV4fl/fl PBS (n = 12), ETV4fl/fl, alb-cre PBS (n = 14), ETV4fl/fl DEN (n = 21), ETV4fl/fl, alb-cre DEN (n = 24). (I-P) Schematic of DEN- and CCL4-induced HCC in WT and ETV4Hep-TG mice (I). The mice were sacrificed at the indicated times, and the livers were harvested. Representative images of the livers of WT and ETV4Hep-TG mice are shown (J). The number of surface tumors and the number of surface tumors larger than 1 mm3 were calculated (K, L). The liver/body weight of the mice was measured (M). The livers were fixed and subjected to H&E staining, and representative images are shown (N). The total number of microscopic tumors per cm2 and the number of microscopic tumors larger than 1 mm2 per cm2 were calculated according to the H&E staining in N (O, P). WT PBS (n = 12), ETV4Hep-TG PBS (n = 17), WT DEN (n = 15), ETV4Hep-TG DEN (n = 16). (Q-T) The DEN-CCL4-induced HCC in ETV4fl/fl and ETV4fl/fl, alb-cre or WT and ETV4Hep-TG mice were sacrificed, the livers of mice were subjected to IHC staining for TNF-α, MAPK11 and CD68. Representative images are shown in Q and S. The intensities of TNF-α, MAPK11 and CD68 staining were quantified in R and T. ETV4fl/fl (n = 3), ETV4fl/fl, alb-cre (n = 3), WT (n = 3), ETV4Hep-TG (n = 3). The column bar graphs were presented as the mean ± SD. In the scatter plots, division line was mean, whiskers were Standard deviation. * P < 0.05, ** P < 0.01, *** P < 0.001, ns, not statistically significant.
Abbreviations: ETV4, ETS translocation variant 4; TNF-α, tumor necrosis factor alpha; MAPK11, mitogen-activated protein kinase 11; DEN, diethylnitrosamine; PBS, phosphate buffered saline; d, days; WT, wild type; Alb-Cre, cre driven by albumin promoter; CCL4, carbon tetrachloride; H&E, hematoxylin and eosin staining; IHC, immunohistochemistry.
A DEN-CCL4 HCC model with ETV4Hep-TG mice was also established (Figure 5I). Total number of surface tumors in the livers of the ETV4Hep-TG mice was higher than those in the WT mice (Figure 5J-K). No significant difference was observed in the number of surface tumors larger than 1 mm3 and the liver/body ratio between the ETV4Hep-TG and WT mice (Figure 5L-M), but total numbers of microscopic tumors and the numbers of large microscopic tumors were robust higher in the ETV4Hep-TG mice than in the WT mice (Figure 5N-P). All these data suggested that ETV4 promoted HCC formation and could be a potential therapeutic target for HCC. In addition, the protein levels of TNF-α, MAPK11 and CD68 were lower in the liver tissues of the ETV4fl/fl, alb-cre mice than in those of the ETV4fl/fl mice (Figure 5Q-R) and significantly higher in the ETV4Hep-TG mice than in the WT mice (Figure 5S-T), suggesting that ETV4 promoted the expression of TNF-α and MAPK11 and hepatic macrophage infiltration during the development of DEN-CCL4-induced HCC.
3.6 ETV4 expression was positively correlated with TNF-α and MAPK11 expression and liver inflammation in human HCC samples
As shown in Figure 1C and Supplementary Figure S1C-F, the level of ETV4 was higher in HCC tumors than in adjacent peritumoral tissues. If ETV4 promoted liver tumorigenesis through TNF-α and MAPK11, there should be higher levels of TNF-α and MAPK11 in tumor tissues than in peritumoral tissues. As expected, there were more TNF-α- and MAPK11-positive cells and higher H-scores of TNF-α and MAPK11 in tumors than in peritumoral tissues according to the IHC staining results of Cohort 2 (Figure 6A-D, Supplementary Figure S6). TNF-α and MAPK11 expression were also higher in tissues with high ETV4 expression than in tissues with low ETV4 expression (Figure 6E). Then the mRNA levels of ETV4, TNF-α and MAPK11 in the HCC tumors in Cohort 3 were analyzed by qRT-PCR. The data showed that ETV4 expression was positively correlated with TNF-α and MAPK11 expression (Figure 6F-G). By analyzing the HCC single-cell transcriptome data from public database. we further found that ETV4 was mainly expressed in tumor cells (Figure 6H), and TNF-α and MAPK11 were co-expressed with ETV4 in tumor cells (Figure 6I-J). We divided the patients in Cohort 2 into different groups according to the levels of TNF-α, MAPK11 and ETV4 and performed Kaplan-Meier survival analysis. The results demonstrated that patients with higher TNF-α level had poorer survival than those with lower TNF-α level (Figure 6K), while the OS had no significant difference between patients with higher MAPK11 and those with lower MAPK11 (Figure 6L). Furthermore, patients with higher levels of ETV4+TNF-α had an even worse survival rate than those with lower levels of ETV4+TNF-α (Figure 6M), while patients with higher levels of ETV4+MAPK11 had poorer survival rate than those with lower levels of ETV4+MAPK11 (Figure 6N). These data suggested that ETV4+TNF-α might be used as prognostic markers for HCC patients. We analyzed the correlation of ETV4 level in tumor with the scores of immune cell types enumerated by xCell, and found that there were more monocytes (Figure 6O) and macrophages (Figure 6P) in ETV4-high tumors than that in ETV4-low tumors, indicating more monocyte and macrophage infiltration in ETV4-high tumors.
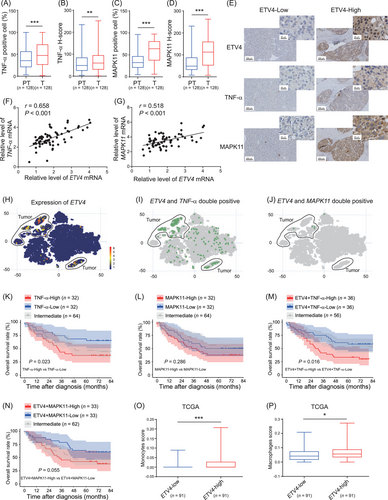
ETV4 expression is significantly correlated with TNF-α and MAPK11 expression in liver tissues of HCC patients. (A-D) A total of 128 paraffin-embedded liver tissues from HCC patients were subjected to IHC staining for TNF-α and MAPK11. The numbers of TNF-α- and MAPK11-positive cells in the peritumoral (PT) and tumoral (T) tissues were calculated using the HALO digital pathology system. The proportion of TNF-α- and MAPK11-positive cells are shown in A and C. The H-scores of TNF-α and MAPK11 are shown in B and D. H-score = weak positive cell percentage × 1+ moderately positive cell percentage × 2+ strongly positive cell percentage × 3. (E) IHC staining of ETV4, TNF-α and MAPK11 were performed in Cohort 2. The samples were subclassified into ETV4-High or ETV4-Low groups according to the average intensity of ETV4, the presentative images of TNF-α and MAPK11 in ETV4-High or -Low groups were shown in E. (F, G) HCC samples from 66 HCC patients (Cohort 3) were collected, and total RNA was extracted and subjected to qRT-PCR to determine the mRNA levels of ETV4, TNF-α and MAPK11. The Pearson correlation coefficient (r) and P value of ETV4 with TNF-α or ETV4 with MAPK11 were analyzed. (H-J) The expression of ETV4 (H) and the double positive of ETV4 with TNF-α (I) or ETV4 with MAPK11 (J) were analyzed based on the scRNA-seq of HCC tumor specimens (https://db.cngb.org/PRHCCdb/). (K-N) Based on the IHC staining for ETV4, MAPK11 and TNF-α from cohort 2, the average intensity of the staining for those proteins was analyzed using a HALO digital pathology system. The samples were classified into high and low groups for TNF-α, MAPK11, ETV4+TNF-α, or ETV4+MAPK11. The Kaplan-Meier overall survival of patients in different groups was analyzed. (O-P) The cell subsets were enumerated by xCell (https://comphealth.ucsf.edu/app/xcell) based on LIHC RNA-seq data in TCGA. The monocytes (O) and macrophage (P) score was shown in ETV4-low and ETV4-high groups. ETV4-low and -high were classified in quartile according to the ETV4 level in TCGA LIHC. In the Box and whiskers plots, boxes represented 25-75% percentile, whiskers were min to max, and division line was median. * P < 0.05, ** P < 0.01, *** P < 0.001.
Abbreviations: ETV4, ETS translocation variant 4; TNF-α, tumor necrosis factor alpha; MAPK11, mitogen-activated protein kinase 11; PT, peritumoral tissue; T, tumor; HCC, hepatocellular carcinoma; scRNA-seq, single cell RNA sequencing; LIHC, liver hepatocellular carcinoma; TCGA, The Cancer Genome Atlas; IHC, immunohistochemistry; qRT-PCR, real-time quantitative PCR.
Taken together, our data suggested that ETV4 promoted liver inflammation and tumorigenesis via enhancing the TNF-α and MAPK11 signaling pathway (Figure 7).
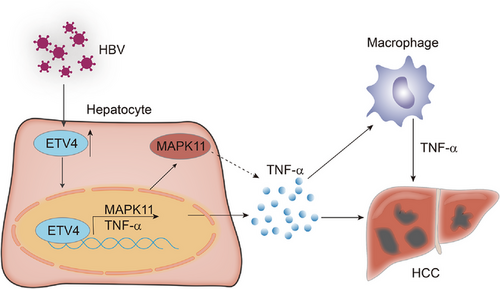
The model of ETV4 promoted liver inflammation and tumorigenesis. ETV4 was upregulated by HBV infection, the upregulation of ETV4 promote the hepatocyte released TNF-α and hepatic TNF-α secretion, thus facilitated macrophages accumulation and promoted tumorigenesis.
Abbreviations: ETV4, ETS translocation variant 4; HBV, hepatitis B virus; TNF-α, tumor necrosis factor alpha; MAPK11, mitogen-activated protein kinase 11.
4 DISCUSSION
In the present study, we found that ETV4 in hepatocytes could promote the inflammation and macrophage accumulation in the liver of mice. Overexpression of ETV4 (ETV4Hep-TG) promoted the HCC formation in mice, while knockout of ETV4 (ETV4fl/fl, alb-cre) inhibited the HCC formation. ETV4 promoted the transcription of TNF-α and MAPK11 both in vitro and in vivo. The ETV4+TNF-α level was negatively correlated with the survival of HCC patients.
As a transcription factor, ETV4 was shown to promote the development of different kinds of cancer by increasing cell proliferation and metastasis. ETV4 directly regulated the transcription of matrix metalloproteinase 13 (MMP13) and promoted the invasion and metastasis of breast cancer [34]. ETV4 also downregulated Cyclin-dependent kinase inhibitor 1A (CDKN1A) expression by binding to the promoter of CDKN1A and increased the proliferation of prostate cancer cells [35]. In addition, ETV4 facilitated Yes-associated protein (YAP)/TEA domain family member (TEAD)-mediated transcriptional activation and enhanced the progression of HCC [36], and regulated the expression of MMP family members to promote tumor metastasis [34, 37, 38]. In this study, we revealed that ETV4 promoted HCC development by regulating liver inflammation via the TNF-α pathway.
TNF-α is an inflammatory cytokine which is abnormally expressed in the liver during damage, and it plays a key role in HCC development. For instance, TNF-α expression was upregulated during DEN- or CCL4-induced acute liver damage [39]. TNF-α expression was enhanced under the conditions of HFD-, choline-deficient and ethionine-supplemented (CDE) diet- or CCL4-induced chronic liver injury [21]. TNF-α expression was also elevated in the livers with hepatic dysfunction, such as cholestasis [20, 40]. Loss of TNFR1 reduced the liver tumorigenesis induced by DEN and HFD [18]. Depletion of TNFR1 inhibited HCC development in MUP-uPA mice, which had high urokinase plasminogen activator (uPA) expression [40]. Our findings revealed that ETV4 promoted the expression of TNF-α and enhanced tumor formation. It is possible that interference of TNF-α or TNFR1 may reduce tumorigenesis induced by ETV4 overexpression.
It had been reported that the inflammatory cytokines IL-6, TNF-α, IL-1β and lymphotoxin-A accelerated the development and aggressiveness of HCC in a mouse model [14-18]. Previous studies had revealed that TNF-α was mainly produced by non-hepatocytes in mdr2-/- mice [20] and was mostly secreted by inflammatory cells during CDE diet-induced liver inflammation [21]. In this study, we surprisingly found that ETV4 promoted the transcription of TNF-α in hepatocytes and led to the higher level of TNF-α in the liver, which induced inflammation and promoted tumorigenesis. It would be worth to further investigate the function of hepatocyte-secreted TNF-α in tumor microenvironment and the transition of chronic inflammation to tumor.
A previous study reported that ETV4 could promote the transcription of C-X-C motif chemokine ligand 1 (CXCL1) and CXCL5 in HCC cells and led to the changes of immune cells in the subcutaneous tumors [36]. However, whether the phenotype obtained from the subcutaneous tumor model would truly occur under the physiological condition needed further investigation. By using orthotopic xenograft models, Xie et al. [41] found that ETV4 upregulated the transcription of programmed cell death-ligand 1 (PD-L1) and C-C motif chemokine ligand 2 (CCL2) and promoted HCC metastasis. Their results may represent the microenvironment in late stage during tumor development since they performed the experiments with the tumor cell line Hepa1-6. In the present study, we found that ETV4 enhanced the inflammation and tumorigenesis by promoting the transcription of TNF-α and MAPK11 in hepatocyte-specific ETV4Hep-TG and ETV4fl/fl, alb-cre mice. These data implied that ETV4 played an important role in inflammation-tumor transition at the early stage of tumor formation, suggesting that ETV4 could affect multiple signaling pathways at different stages of HCC development.
HBV is one of the risk factors that induces chronic hepatitis. Other types of hepatitis are observed in the clinic, such as NASH, alcoholic steatohepatitis and cholestasis-induced hepatitis [2, 4]. The present study indicated that HBV induced the expression of ETV4, which promoted liver tumorigenesis. In addition, we found that ETV4 was upregulated in alcohol assumption- and non-alcoholic fatty liver disease-associated HCC compared to normal livers. We also observed that the level of ETV4 mRNA was increased in the livers of mice treated with DEN or CCL4. These data indicated that inflammation could also trigger the transcription of ETV4. It suggested that inflammation and ETV4 formed positive feedback. If so, it is possible that ETV4 also plays a critical role during the development of inflammation-related liver diseases other than HCC.
Previous studies showed that the monocyte-derived macrophages played an important role during the HCC development [42-44]. Our data showed that there was more macrophage accumulation in ETV4Hep-TG mice. However, we did not examine whether the macrophage enhancement in ETV4Hep-TG mice was from monocyte differentiation or Kupffer cell proliferation, which needed further investigation. In addition, the functions of different subpopulations of macrophages were divergent during HCC development [22, 45, 46]. Therefore, it is worthwhile to further explore which subpopulation of macrophage is affected by ETV4 to better understand the function of ETV4 in inflammation and HCC formation. We also observed more neutrophil accumulation in ETV4Hep-TG mice. It has been reported that N2 neutrophils promoted tumor formation by enhancing reactive oxygen species [47]. It will be worth to study what type of neutrophils was accumulated in ETV4Hep-TG mice in the future.
5 CONCLUSIONS
In conclusion, we found that ETV4 was highly expressed in HCC, and its expression was negatively associated with the survival of HCC patients. ETV4 promoted liver inflammation and enhanced liver tumorigenicity in vivo. Mechanistically, ETV4 could promote the transcription of TNF-α and MAPK11 directly, and enhanced liver inflammation by increasing macrophages and neutrophils. We demonstrated that the level of ETV4+TNF-α was negatively associated with the survival of HCC patients, which may be potential prognostic markers for HCC patients.
DECLARATIONS
AUTHOR CONTRIBUTIONS
Dandan Qi and Min Lu designed and performed the experiments, and wrote the draft. Pengfei Xu performed the IHC staining. Xinli Yao, Yongchen Chen, Yahua Cui and Lipeng Gan supported the mouse experiments. Yong Li performed the statistical analysis. Xiaomei Tong and Shuhong Liu contributed to the constructive discussions. Jingmin Zhao provided the clinical samples and contributed to the constructive discussions. Ningning Liu interpreted data and revised the manuscript. Xin Ye supervised the study and wrote the manuscript.
ACKNOWLEDGMENTS
We are grateful to Xiaolan Zhang, Meng Wan and Xiaopeng Yang for providing technical support for the IHC staining analysis. We are also grateful to Nanjing Freethinking Biotechnology Co., Ltd. (China) for providing HALO pathology analysis services.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
CONSENT FOR PUBLICATION
Not applicable.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The HCC tissues in Cohorts 1-3 were all from the Fifth Medical Center of Chinese PLA General Hospital. This study was approved by the Ethics Committee of the Fifth Medical Center of Chinese PLA General Hospital (GH-7-20180563). All the experiments were performed according to approved guidelines and regulations. Animal studies were approved by Institute of Microbiology, Chinese Academy of Sciences, Research Ethics Committee and were performed according to the application guidelines and regulations (APIMCAS2019009).
Open Research
DATA AVAILABILITY STATEMENT
The overall survival of ETV4 in TCGA was from GEPIA (http://gepia.cancer-pku.cn/). The gene expression profiles were from TCGA datasets (https://www.cancer.gov/ccg/research/genome-sequencing/tcga), the Gene Ommnibus database (GEO) was form NCBI (GEO DataSets - NCBI (nih.gov)). The immune cell enrichment profiles were from xCell (https://xcell.ucsf.edu/). The quantitative form of IHC staining in Cohort 2 by HALO digital platform are available from the corresponding author for reasonable request.




