HMMR alleviates endoplasmic reticulum stress by promoting autophagolysosomal activity during endoplasmic reticulum stress-driven hepatocellular carcinoma progression
Abstract
Background
The mechanism of hepatitis B virus (HBV)-induced carcinogenesis remains an area of interest. The accumulation of hepatitis B surface antigen in the endoplasmic reticulum (ER) of hepatocytes stimulates persistent ER stress. Activity of the unfolded protein response (UPR) pathway of ER stress may play an important role in inflammatory cancer transformation. How the protective UPR pathway is hijacked by cells as a tool for malignant transformation in HBV-related hepatocellular carcinoma (HCC) is still unclear. Here, we aimed to define the key molecule hyaluronan-mediated motility receptor (HMMR) in this process and explore its role under ER stress in HCC development.
Methods
An HBV-transgenic mouse model was used to characterize the pathological changes during the tumor progression. Proteomics and transcriptomics analyses were performed to identify the potential key molecule, screen the E3 ligase, and define the activation pathway. Quantitative real-time PCR and Western blotting were conducted to detect the expression of genes in tissues and cell lines. Luciferase reporter assay, chromatin immunoprecipitation, coimmunoprecipitation, immunoprecipitation, and immunofluorescence were employed to investigate the molecular mechanisms of HMMR under ER stress. Immunohistochemistry was used to clarify the expression patterns of HMMR and related molecules in human tissues.
Results
We found sustained activation of ER stress in the HBV-transgenic mouse model of hepatitis-fibrosis-HCC. HMMR was transcribed by c/EBP homologous protein (CHOP) and degraded by tripartite motif containing 29 (TRIM29) after ubiquitination under ER stress, which caused the inconsistent expression of mRNA and protein. Dynamic expression of TRIM29 in the HCC progression regulated the dynamic expression of HMMR. HMMR could alleviate ER stress by increasing autophagic lysosome activity. The negative correlation between HMMR and ER stress, positive correlation between HMMR and autophagy, and negative correlation between ER stress and autophagy were verified in human tissues.
Conclusions
This study identified the complicated role of HMMR in autophagy and ER stress, that HMMR controls the intensity of ER stress by regulating autophagy in HCC progression, which could be a novel explanation for HBV-related carcinogenesis.
List of abbreviations
-
- 4E-BP1
-
- eukaryotic translation initiation factor 4E-binding protein 1
-
- ALT
-
- alanine aminotransferase
-
- AST
-
- aspartate aminotransferase
-
- ATF4
-
- activating transcription factor 4
-
- ATF6
-
- activating transcription factor 6
-
- Baf A1
-
- bafilomycin A1
-
- CHOP
-
- c/EBP homologous protein
-
- CHX
-
- cycloheximide
-
- CQ
-
- chloroquine
-
- CTSD
-
- cathepsin D
-
- DMSO
-
- dimethyl sulfoxide
-
- EIF2S1
-
- eukaryotic translation initiation factor 2 subunit alpha
-
- ER
-
- endoplasmic reticulum
-
- GCN2
-
- general control nonderepressible 2
-
- GRP78
-
- glucose-regulated protein78
-
- GGH
-
- ground-glass hepatocyte
-
- HBsAg
-
- hepatitis B surface antigen
-
- HBV
-
- hepatitis B virus
-
- HCC
-
- hepatocellular carcinoma
-
- HMMR
-
- hyaluronan-mediated motility receptor
-
- IRE1α
-
- inositol-requiring enzyme-1
-
- LAMP1
-
- lysosome-associated membrane protein 1
-
- LC3
-
- microtubule associated protein 1 light chain 3
-
- MITF
-
- microphthalmia-associated transcription factor
-
- mTOR
-
- mechanistic target of rapamycin kinase
-
- P62/SQSTM1
-
- sequestosome 1
-
- PERK
-
- protein kinase R-like endoplasmic reticulum kinase
-
- PPP3CB
-
- protein phosphatase 3 catalytic subunit beta
-
- qRT-PCR
-
- quantitative real-time polymerase chain reaction
-
- RBX1
-
- ring-box 1
-
- SKP1
-
- S-phase kinase associated protein 1
-
- TFEB
-
- transcription factor EB
-
- TM
-
- tunicamycin
-
- TRIM29
-
- tripartite motif containing 29
-
- TUDCA
-
- tauroursodeoxycholate
-
- Ub
-
- ubiquitin
-
- UPR
-
- unfolded protein response.
-
- XBP1
-
- X-box binding protein 1
-
- siRNA
-
- small interfering RNA
-
- IP
-
- immunoprecipitation
-
- Co-IP
-
- coimmunoprecipitation
-
- SDS-PAGE
-
- Sodium dodecyl-sulfate polyacrylamide gel electrophoresis
-
- RNA-seq
-
- RNA sequencing
1 BACKGROUND
Hepatocellular carcinoma (HCC) is the most common type of liver cancer and accounts for approximately 80% of liver cancers [1, 2]. Chronic hepatitis B virus (HBV) infection results in an aggressive disease course leading to HCC [3, 4]. Currently, anti-HBV therapy can only reduce viral load but does not eliminate the virus [5]. Therefore, exploring the pathogenesis of HBV-associated HCC to find new therapeutic strategies remains an important issue. The oncogenic mechanisms of HBV related to the integration of HBV DNA into the host chromosomes and HBV protein X-induced hepatocarcinogenesis are well documented [3, 6, 7]. HBV DNA replication is error prone, leading to naturally occurring nucleotide mutations in all HBV-encoding genes [8]. The mutations found in chronic HBV carriers are associated with a high risk of HCC [9, 10]. Mutation of HBV surface antigen (HBsAg) results in aberrant accumulation of HBsAg in the endoplasmic reticulum (ER), which is a pathologic feature of ground-glass hepatocytes (GGHs) [11]. This phenomenon was verified in transgenic mouse models that overproduce large HBsAg in hepatocytes, displaying GGH features and developing hepatocellular neoplasia [12-14]. Several studies have reported that the retention of HBsAg in the ER induces ER stress and activates the unfolded protein response (UPR) to promote HCC progression [10, 14]. However, the intrinsic regulation of UPR signaling in the process of inflammation-fibrosis-hepatocarcinogenesis driven by chronic ER stress remains unclear.
ER stress induced by the overload of synthetic proteins in the ER lumen can activate three classical UPR pathways, protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK), inositol-requiring enzyme 1 (IRE1α), and activating transcription factor 6 (ATF6). UPR signal transduction can enhance ER protein folding capacity, degrade synthesized proteins by ER-associated degradation (ERAD), and reduce overall protein synthesis to alleviate this stress or execute cell death, which is defined as “adaptive UPR” and “terminal UPR” [15-17]. To this end, both “adaptive UPR” and “terminal UPR” serve as protectors of cellular homeostasis and eliminate aberrant cells. However, activation of the UPR pathways by stimuli is a double-edged sword in the process of tumorigenesis depending on acute ER stress and chronic ER stress. During tumorigenesis, how “adaptive UPR” rather than “terminal UPR” is hijacked by cells upon persistent stimuli-induced chronic ER stress is unknown.
The three pathways of the UPR can regulate cell survival by modulating multiple downstream molecules to induce autophagy [18]. The activation of ER stress associated with HBsAg can also induce autophagy [18, 19]. UPR signaling regulates viral replication by regulating autophagy [20], since viruses need the host ER to produce increased quantities of viral proteins to continue replication. Autophagic degradation of misfolded proteins can be used as compensation to ERAD, with the function of reducing ER stress and inhibiting cell death [19-21]. This finding suggests that virus-associated UPR signaling induces cellular autophagy as a major protein degradation mechanism to alleviate ER stress and cell survival [22]. It is unclear whether chronic ER stress or the attenuated ER stress response contributes to HCC development.
Hyaluronan mediated motility receptor (HMMR), a receptor for hyaluronic acid, plays an important role in cell migration, growth, and differentiation [23]. As a spindle assembly factor, HMMR regulates the motor activity of dynein and is involved in the regulation of cell division and the cell cycle [24, 25]. Most studies reported that HMMR functions as a tumorigenicity gene [26] or as a pro-cancer modifier [27], and the expression of HMMR was elevated in various tumors and was associated with poor prognosis [28, 29]. However, in a mouse model of pancreatic cancer, deletion of HMMR appears to act as a pro-cancer phenotype [30]. Loss of function of HMMR is the initiating event in the development of seminoma [31]. Moreover, the function of HMMR in the HBV-related hepatitis-fibrosis-HCC process has not been fully investigated.
In this study, we sought to elucidate the molecular mechanism that regulated the expression of the key gene HMMR screened in this process and its significance for HCC progression under ER stress.
2 MATERIALS AND METHODS
2.1 Antibodies and reagents
Primary antibodies used for Western blotting include human HMMR (1:1000; ab185728, Abcam, Cambridge, Cambridgeshire, UK), mouse HMMR (1:1000; sc-515221, Santa Cruz, Dallas, TX, USA), human PERK (1:500; 24390-1-AP, Proteintech, Wuhan, Hubei, China), mouse PERK (1:500; 3192s, CST, Boston, MA, USA), mouse p-PERK (1:500; orb336657, Biorbyt Limited, Cambridge, Cambridgeshire, UK), human c/EBP homologous protein(CHOP; 1:1000; 15204-1-AP, Proteintech), mouse CHOP (1:1000; 2895t, CST), mouse and human GRP78 (1:1000; 11587-1-AP, Proteintech), α-tubulin (1:3000; 66031-1-Ig, Proteintech), β-actin (1:3000; M1210-2, HUABio, Wuhan, Hubei, China), S-phase kinase associated protein 1(SKP1; 1:1000; 10990-2-AP, Proteintech), ring-box 1 (RBX1; 1:1000; 14895-1-AP, Proteintech), TRIM29 (1:1000; 17542-1-AP, Proteintech), Flag Tag (1:1000; M1403-2, HUABio), MYC Tag (1:1000; 16286-1-AP, Proteintech), LC3 (1:1000; 12741s, CST), P62 (1:1000; p0067, Sigma, Aldrich Chemie, Taufkirchen, Germany), CTSD (1:1000; 21327-1-AP, Proteintech), LAMP1 (1:1000; ab24170, Abcam), TFEB (1:1000; 13372-1-AP, Proteintech), MITF (1:1000; 13092-1-AP, Proteintech), lamin B1 (1:1000; 12987-1-AP, Proteintech), 4EBP1 (1:1000; 9644t, CST), p-4EBP1 (1:1000; 2855t, CST), mTOR (1:1000; 2983s, CST), p-mTOR (1:1000; 5536s, CST), PPP3CB (1:1000; 55148-1-AP, Proteintech), and Ub (1:1000; sc-8017, Santa Cruz). Antibodies were used for immunoprecipitation (IP) assays, including HMMR (1:10; ab108339, Abcam), Flag Tag (1:20; M1403-2, HUABio), and CHOP (1:100, 2895T, CST). Primary antibodies were used for immunofluorescence, including HMMR (1:200; sc-515222, Santa Cruz) and TRIM29 (1:200; PA5-102978, Invitrogen, Carlsbad, CA, USA). Primary antibodies were used for immunohistochemistry (IHC) staining, including HMMR (1:250; ab124729, Abcam), TRIM29 (1:200; PA5-102978, Invitrogen), GRP78 (1:200; 11587-1-AP, Proteintech), P62 (1:200; 18420-1-AP, Proteintech), and Beclin1 (1:200; 11306-1-AP, Proteintech).
Cells were treated with 10 μmol/L MG132 (C2211, Sigma), 100 μg/mL cycloheximide (CHX; 239763-M, Sigma), 50 μmol/L chloroquine (CQ; C6628, Sigma), 5 μg/mL tunicamycin (TM; HY-A0098, MedChemExpress, Shanghai, China), 100 μmol/L tauroursodeoxycholate (TUDCA; HY-19696, MedChemExpress) or 50 nmol/L bafilomycin A1 (Baf A1; ab120497, Abcam). The reagents were dissolved in dimethyl sulfoxide (DMSO; D2650, Sigma) and further diluted in cell culture medium. To control the DMSO-induced cytotoxicity, same percentage of DMSO was taken as a control.
2.2 Animals
HBV-transgenic (tg) mice [C57BL/6J-Tg(Alb1HBV)44Bri/J] were purchased from Vital River Laboratories (002226, Beijing, China). The mice were maintained in specific pathogen free carriers with free access to water and food [12]. The HBV large envelope polypeptide sequence was inserted downstream of the mouse albumin promoter, thus inducing hepatocyte-specific overexpression of the large envelope polypeptide in HBV-tg mice. HBV-tg and control mice were maintained for 4, 6, 8, 10, 12, 14, 16, and 18 months. Mice were euthanized by overdose anesthesia, and tissue samples were collected at the designated endpoint. Animal studies were approved by the Animal Care and Use Committee of National Translational Science Centre for Molecular Medicine (2020-NTSCMM-ID004).
2.3 Patient tissue samples
Forty-seven pairs of HCC liver tissue for IHC to detect the expression of HMMR; another 25 hepatitis liver tissues and 25 HCC liver tissues for IHC to detect the expression of HMMR, TRIM29, and the UPR and autophagy-related genes; 29 pairs of HCC liver tissue for Quantitative real-time PCR (qRT-PCR); and 5 pairs of HCC liver tissue for Western blotting were obtained from Xijing Hospital of Fourth Military Medical University (Xi'an, Shaanxi, China). Hepatitis liver tissues were obtained by puncture biopsy of patients with HBV, while HCC tissues were obtained from surgically resected specimens of patients with HBV-related HCC. All patients had a definite diagnosis of HBV and all the sections were reviewed by a pathologist. Ethical approval was obtained from the Ethics Committee of the Fourth Military Medical University (202003-216).
2.4 HCC patient data and processing
RNA sequencing (RNA-seq) data of HCC patient samples (including 374 liver tumor tissues and 50 adjacent tissues) were downloaded from TCGA (https://portal.gdc.cancer.gov/). The mRNA expression values of HMMR gene for TCGA_LIHC were subjected to log2(FPKM+1) transformation. HMMR mRNA expression data of human hepatitis, cirrhosis, and HCC tissues in GSE83148, GSE25097, and GSE14520 were downloaded from Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/). The “Limma” package in R software (version 4.1.0; Auckland University, Auckland, New Zealand) was used to identify differentially expressed genes (DEGs). P < 0.05 and a fold-change (FC) > 1.0 were considered to indicate a statistically significant difference.
2.5 Cell culture and transfection
The human liver cancer cell lines MHCC-97H and HCCLM3 and the human embryonic kidney cell line HEK-293T were purchased from the Shanghai Institutes for Biological Sciences (Shanghai, China). Cells were cultured in DMEM (11965092, Invitrogen) with 10% fetal bovine serum (10100147, Invitrogen), 100 U/mL penicillin, and 100 μg/mL streptomycin (15240062, Invitrogen). The small interfering RNA (siRNAs) targeting HMMR and TRIM29 were designed and synthesized by GenePharma Co., Ltd. (Suzhou, Jiangsu, China), and the siRNA sequences are listed in Supplementary Table S1. HMMR overexpression lentivirus and the vector were purchased from Genechem Technology (Shanghai, China). The plasmid expressing ATF6 and the vector control plasmid were constructed by Dr. Ying Liu (Institute of Basic Medical Sciences Chinese Academy of Medical Sciences, Beijing, China). The plasmid pcDNA3.1-TRIM29 and the vector control plasmid pcDNA3.1 were purchased from Tsingke Co., Ltd (Beijing, China). The plasmids expressing activating transcription factor 4 (ATF4), X-box binding protein 1s (XBP1s), HMMR promoter reporter (-2061/+61), and HMMR promoter-truncated reporter (-933/+61) and the corresponding vector plasmids were purchased from Genechem Co., Ltd. The plasmids expressing HMMR-N-Flag and CHOP and vectors were purchased from Sino Biological (Beijing, China). The dual-labelled mCherry-GFP-LC3 plasmid was purchased from Beyotime (D2816, Shanghai, China). All transfections were performed according to the instructions using Lipofectamine 2000 (11668500, Invitrogen).
2.6 Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was performed using a ChIP Kit (ab500, Abcam). Briefly, the samples were fixed and shattered by ultrasound. The 200-1000 bp DNA fragments were incubated with beads and 5 μg IgG or CHOP antibodies. After DNA purification of the samples, qRT-PCR reactions generated products from the promoter region of the HMMR gene. Primers based on the predicted binding sites were used: 5'-CTGCTTGACCACTCCACAAA-3' (forward primer) and 5'-GCTCCTGTACGGAAGCGTAA-3' (reverse primer).
2.7 IP assay and Western blotting
The interaction of HMMR and TRIM29 in MHCC-97H and HCCLM3 cells was detected using a coimmunoprecipitation (Co-IP) kit (26149, Pierce, Rockford, IL, USA) according to the manufacturer's instructions. Endogenous and exogenous HMMR and ubiquitination were detected using an IP Kit according to the manufacturer's instructions (26147, Pierce). IP and Co-IP were assayed via Western blotting. Western blotting analysis was performed according to a standard protocol. Briefly, total cell lysates or coprecipitates were electrophoresed by Sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to PVDF membranes (Millipore, Billerica, MA, USA). The membranes were blocked with 5% skim milk powder for 1 hour and then incubated with primary antibodies at 4°C overnight. The system was reacted with anti-mouse IgG (31430, Pierce) or anti-rabbit IgG (31460, Pierce) secondary antibody at room temperature for 1 hour. The reactive bands were visualized by a ChemiDocTM Touch Imaging System (Bio-Rad, Hercules, CA, USA).
2.8 Quantitative real-time PCR
Total RNA was extracted from liver tissue of mice or cultured cells using the Total RNA Kit II (D6934-01, Omega Scientific, Tarzana, CA, USA) and reverse-transcribed into complementary DNA by the PrimeScript RT reagent kit (RR036A, Takara Bio, Yokkaichi, Japan). A TBGreen Premix ExTaq kit (TaKaRa Bio, RR820A) was used to amplify the single-stranded complementary DNA with the Stratagene Mx3005P Real-Time PCR System (Agilent Technologies, Shanghai, China). The ΔΔCt method was used for computation of relative mRNA concentrations, and data were normalized to GAPDH expression. The primers are listed in the Supplementary Table S2.
2.9 Immunofluorescence
HCC cells were seeded in 35 mm confocal dish. After overnight culture and treated with TM for 8 hours, cells were fixed in 4% paraformaldehyde for 20 minutes, then washed with PBS. 0.2%Triton X-100 was used to perforate and 5% goat serum were used to block non-specific binding. Cells were incubated with anti-HMMR and anti-TRIM29 antibodies at 4°C overnight. On the next day, cells were washed with PBS, and incubated with the Alexa Fluor 488 anti-mouse IgG (1:200; Invitrogen) and Alexa Fluor 555 anti-rabbit IgG (1:200; Invitrogen) at room temperature for 1 hour. 4,6-diamidino-2-phenylindole (DAPI, Beyotime) was used for nuclei staining. Images were captured by a confocal fluorescence microscopy (A1R Confocal; Nikon, Tokyo, Japan).
2.10 Alanine aminotransferase (ALT) and aspartate aminotransferase (AST) analysis
Serum ALT and AST were measured by assay kits according to the manufacturer's instructions (C009-2 and C010-2, Nanjing JianCheng Bioengineering Institute, Nanjing, China). Briefly, experiments were performed in 96-well plates. The 5 μL mouse serum was added to the wells and reacted with pre-warmed ALT/AST matrix solution at 37°C for 30 minutes, and 5 μL serum were added to the control wells and reacted with pre-warmed 2,4-dinitrophenylhydrazine solution at 37°C for 20 minutes. Then 200 μL sodium hydroxide solution (0.4 mol/L) was added to all the wells. The OD value of each well was measured at 510 nm using a microplate reader (Microplate Reader 550, Bio-Rad). To obtain the absolute OD value, the OD value of the experimental well was subtracted from the OD value of the control well. The corresponding ALT/AST activity values were obtained by checking the standard curve.
2.11 IHC staining
IHC staining was performed using a streptavidin-peroxidase kit according to the manufacturer's instructions (SP-9000, Zhongshan Jinqiao Co., Beijing, China). Paraffin sections were baked overnight at 50°C-60°C and then routinely dewaxed and hydrated. Antigen retrieval was performed according to the primary antibody instructions. The sections were incubated with goat serum to block nonspecific binding. Subsequently, they were incubated with primary antibody at 4°C overnight, then incubated with HRP-conjugated secondary antibody. Samples were visualized with a DAB kit (ZLI-9019, Zhongshan Jinqiao Co.) and counterstained with hematoxylin (G1140, Solarbio Life Sciences, Beijing, China). After dehydration and sealing, sections were observed under a microscope. The slides were scanned by an automatic digital slide scanner (3DHistech Ltd, Budapest, Hungary, Pannoramic MIDI II). Quantitative analysis was performed with HALO™ Image Analysis software, and the data are shown as the average cytoplasmic OD.
2.12 Hematoxylin and eosin (H&E) and sirius red staining
The 4% paraformaldehyde-fixed liver tissue from mice was paraffin-embedded and sliced. H&E staining was performed using standard methodology. Sirius red staining was performed as previously described [32]. Briefly, for H&E staining, paraffin sections were baked for 2 hours at 50°C-60°C and then routinely dewaxed and hydrated, stained with hematoxylin (G1140, Solarbio Life Sciences) for 3 minutes. After the sections were stained with eosin solution (Zhuhai Beso Bio Co., Ltd., Zhuhai, China) for 2 minutes, and routinely dehydrated, cleaned, and mounted. For sirius red staining, the sections were treated by same steps above instead of staining with 0.1% sirius red solution (365548, Sigma) for 1 hour.
2.13 LysoTracker Red staining
LysoTracker Red (L8010, Solarbio Life Sciences) was diluted in DMEM at a ratio of 1:20,000 and incubated at 37°C for 2 hours in the dark. The area and intensity of red fluorescence indicated the lysosomal activity.
2.14 Transmission electron microscopy
For transmission electron microscopy, freshly dissected liver tissue or cell samples were fixed in 2.5% glutaraldehyde (Fuchen Chemical Reagent Co., Ltd, Tianjin, China) for 24 hours, subjected to ultrathin sectioning, and examined with electron microscopy (JEOL, Tokyo, Japan, JEM-1230). Five micrographs were captured by electron microscopy, and the number of lysosomes per cell was counted.
2.15 Dual-luciferase reporter assay
The transcription of HMMR regulated by CHOP was detected by the Dual-Luciferase Reporter Assay System (E1980, Promega, Shanghai, China). A total of 1 μg CHOP expression plasmid, 200 ng HMMR luciferase reporter plasmid, and 10 ng Renilla luciferase control reporter plasmid (pRL-TK) were cotransfected into HEK293T cells in a 24-well plate. Cells were tested using Luciferase Assay System (E5311, Promega) after 24 hours transfection.
2.16 CCK8 assay
The ability of cell proliferation was assessed using CCK8 (C0005, Topscience, Shanghai, China) according to the manufacturer's instructions. Cells were seeded in 96-well plates with a cell density of 3,000 cells per well. After treated with TM (5 μg/L) and/or CQ (50 μmol/L) for 48 hours, the absorbance of each well was measured by a microplate reader at an excitation wavelength of 450 nm.
2.17 RNA-seq and gene set enrichment analysis
The 4-, 10- and 16-month-old HBV-tg mice and age-matched control mice were euthanized (n = 4). Total RNA from liver tissue was collected for RNA-seq analysis (Novogene, Beijing, China). Moreover, total RNA from MHCC-97H cells stably transfected with HMMR or the vector was subjected to RNA-seq (BGI gene, Shenzhen, China). Briefly, RNA samples were denatured at suitable temperature to open their secondary structure, and mRNA was enriched by oligo(dT)-attached magnetic beads. After enrichment, mRNA was fragmented using fragmentation buffer and reverse transcribed into cDNA. Double-stranded cDNA fragments were synthesized and subjected to end-repair and PCR amplified. Then single-stranded PCR products were produced via denaturation and single-stranded cyclized products were produced. Single-stranded circle DNA molecules were replicated via rolling cycle amplification, and a DNA nanoball (DNB) which contained multiple copies of DNA was generated. Sufficient quality DNBs were then loaded into patterned nanoarrays using high-intensity DNA nanochip technique and sequenced through combinatorial Probe-Anchor Synthesis. The sequencing data were collected and gene set enrichment analysis (GSEA) was performed using GSEA version 3.0 (http://software.broadinstitute.org/gsea/downloads.jsp, the Broad Institute of MIT and Harvard) to investigate the biological characteristics of HMMR overexpression.
2.18 Tandem mass tag proteomics and gene ontology analysis
The 4-, 10- and 16-month-old HBV-tg mice and age control mice were euthanized (n = 3), and liver tissue was collected for tandem mass tag proteomics (Applied Protein Technology, Shanghai, China). For mass spectrometry with tandem mass tags, mouse liver tissue was lysed by SDT (4% (w/v) SDS, 100 mmol/L Tris/HCl pH 7.6, 0.1 mol/L DTT) using the filter-aided proteome preparation method and trypsinized. The resultant peptides were labelled with tandem mass tag reagents (Invitrogen). Samples were fractionated and ionized on a HPLC system (Easy nLC) and detected by a mass spectrometer (Q-Exactive). Raw files were processed by Mascot2.2 and Proteome Discoverer1.4 software (Thermo Fisher Scientific, Waltham, MA, USA) for inventory identification and quantitative analysis. The data was collected and the Gene Ontology (GO) analysis was done using the phyper function in R software (version 3.6.0; Auckland University).
2.19 Shotgun mass spectrometric
For Shotgun Protein Spectrum (Applied Protein Technology), the Co-IP solution was lysed and enzymatically digested. Briefly, dithiothreitol was added to the samples and mixed at 37°C for 1.5 hours. Then iodoacetamide was added into the mixture to block reduced cysteine residues and the samples were incubated for 30 minutes in darkness. Next, the samples were transferred to the filters. The filters were washed with 100 μL UA buffer (8 mol/L Urea, 150 mmol/L Tris-HCl, pH 8.0) three times and 100 μL 25 mmol/L NH4HCO3 buffer twice. Finally, trypsin was added to the samples and incubated at 37°C for 15-18 hours, and the resulting peptides were collected as a filtrate. The peptides were desalted on C18 Cartridges (Empore SPE Cartridges C18 (standard density), bed I.D. 7 mm, volume 3 mL, Sigma), concentrated by vacuum centrifugation, and reconstituted in 40 μL of 0.1% (v/v) formic acid. The lysate was isolated using HPLC system (Easy-nLC). The separated peptides were analyzed using Q-Exactive mass spectrometer (Thermo Fisher Scientific). Raw data were processed and searched by MaxQuant software (Max Planck Institute of Biochemistry, Martinsried, Germany).
2.20 Protein-protein interaction analysis
Time-series analysis was performed using the Short Time-series Expression Miner (STEM, version 1.3.11; http://www.cs.cmu.edu/~jernst/stem/). The protein-protein interaction (PPI) network was analyzed using the STRING database (https://string-db.org/) for Mus musculus. STRING was used to analyze PPI networks of 181 continuously upregulated proteins at three stages with a combined confidence score > 0.400 as the cut-off criteria. CytoHubba (version 0.1, http://apps.cytoscape.org/download/stats/CytoHubba/) is a Cytoscape (version 3.7.0, https://cytoscape.org/) plugin for analyzing the centrality of protein interaction networks that are representative of potentially crucial proteins in the network [33]. The values of centrality parameters were sorted to obtain the top 8 ranked proteins. For exploration of key hubs of networks, maximal clique centrality (MCC), maximum neighborhood component (MNC) centrality, degree centrality, eccentricity centrality, closeness centrality, and radiality centrality were identified, and proteins with high consistency scores from six different algorithms were defined as candidate crucial proteins.
2.21 Statistical analysis
The experiments were carried out with no less than three biological replicates or three independent repeats, and the data were analyzed as the means ± standard error of mean (SEM) using GraphPad Prism v8.01 software (GraphPad Software Inc., San Diego, CA, USA). Significance was tested with Student's t test, Welch's t test, one-way ANOVA, or two-way ANOVA. Correlations were evaluated through Spearman test. P < 0.05 was considered statistically significant.
3 RESULTS
3.1 HBV-tg mouse model with the hepatitis-liver fibrosis-HCC progression displayed chronic and attenuated ER stress
To observe the pathology of the natural disease course with hepatitis-fibrosis-HCC progression in vivo, HBV-tg mice overexpressing HBV large surface protein (LHB) in the liver were maintained and euthanized at 4, 6, 8, 10, 12, 14, 16, and 18 months. Liver tissues and sera were collected for H&E staining, sirius red staining, and ALT/AST examination. H&E staining of liver tissues indicated that both male and female HBV-tg mice displayed inflammatory cell infiltration and pathological changes compared to age-matched control mice, and generally, the abnormalities in the male HBV-tg mice were more severe and appeared earlier than those in the female HBV-tg mice (Supplementary Figure S1A). The male HBV-tg mice obviously displayed vacuole-like changes in the cytoplasm and increased heterogeneity in the nucleus early at 10 months, while the female HBV-tg mice displayed changes later at 16 months (Supplementary Figure S1A). Compared with that of the age-matched control mice, sirius red staining showed that the total collagen content in the male HBV-tg mice was significantly enhanced early at 8 months (P < 0.001), while it was observed in the female HBV-tg mice at 10 months (P < 0.010) (Supplementary Figure S1B). ALT and AST analyses demonstrated that the HBV-tg mice displayed severe liver injury throughout the course of liver disease, which was more serious in the male HBV-tg mice (Supplementary Figure S1C). Along with node development, the relative liver-to-body weight ratio of the male HBV-tg mice was significantly increased at 14-18 months (Supplementary Figure S2A). We summarized the overall information of liver nodes in male and female HBV-tg mice and found that nodes on the liver surface of the male HBV-tg mice developed earlier and deteriorated (Supplementary Table S3, Supplementary Figure S2B). Typical ground-glass cells were observed in the livers of 16-month-old male mice (Figure 1A). We then performed transmission electron microscopy in the liver tissues of 16-month-old HBV-tg mice to investigate the pathological effects of LHB on organelle ultrastructure. We observed severe ultrastructural alterations of the ER in HBV-tg mouse hepatocytes (Figure 1B). Compared with the neatly and regularly arranged ER of the control mice, the ER of the HBV mice appeared to be particularly disorganized, displaying ER dilation, vesiculation, and fragmentation (Figure 1B), indicative of severe ER stress. We then defined 4-month-old male HBV-tg mice as the hepatitis group, 10-month-old male HBV-tg mice as the liver fibrosis group, and 16-month-old male HBV-tg mice as the HCC group. We performed tandem mass tag proteomics in hepatitis, fibrosis, and HCC group mice and the age-matched control male mice (n = 3). The results showed that there were 214, 271, and 450 significantly differentially upregulated proteins and 172, 193, and 527 significantly differentially downregulated proteins in the 4-, 10-, and 16-month-old mice, respectively. To further study the pathogenic mechanism of HCC progression in HBV-tg mice, we performed GO enrichment analysis on the union of these differentially expressed proteins, which showed that the ER was the most common category and that the categories of ER stress, unfolded proteins, and misfolded proteins were significantly enriched (Figure 1C). Data from qRT-PCR and Western blotting indicated that ER stress-related markers were continuously activated in the liver tissue from the HBV-tg mice with hepatitis-fibrosis-HCC progression (Figure 1D-E). We found that the intensity of ER stress was more severe in the early stages of the disease process than in the stage of HCC by normalizing GRP78 levels of HBV-tg to the same age control groups (Figure 1F). These results indicated that HBV-tg mice displayed the characteristics of clinical HCC development in an age- and sex-dependent manner and represented an \in vivo model of ER stress.
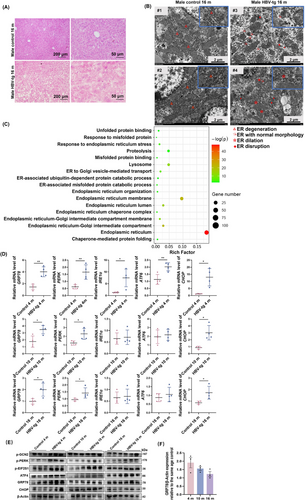
The HBV-tg mouse model with hepatitis-liver fibrosis-HCC progression displayed chronic ER stress. (A) H&E staining of liver tissue from 16-month-old HBV-tg mice and control mice of the same age, which showed ground-glass cells. (B) Ultrastructural alterations in the hepatocytes of the 16-month-old HBV-tg mice compared with the control mice of the same age revealed by transmission electron microscope. (C) Gene ontology analysis of differentially expressed proteins identified by mass spectrometry in liver tissue of 4-, 10- and 16-month-old HBV-tg mice vs. control mice of the same age. (D) The mRNA expression of UPR-related markers determined by qRT-PCR in liver tissue of 4-, 10- and 16-month-old HBV-tg mice and control mice of the same age (n = 4). (E) Western blotting analysis of the expression of UPR-related proteins in liver tissue of the HBV-tg mice and control mice of the same age (n = 3). (F) Protein levels of GRP78 relative to the age-matched control group (normalized to β-actin, n = 3). Two-tailed Student's t test or Welch's t test was used to test the significance of differences between two groups; data are represented as the mean ± SEM. *, P < 0.05; **, P < 0.01.
Abbreviations: H&E, hematoxylin-eosin; ATF4, activating transcription factor 4; ATF6, activating transcription factor 6; CHOP, C/EBP homologous protein; ER, endoplasmic reticulum; GRP78, glucose-regulated protein78; HBV, hepatitis B virus; PERK, protein kinase R-like endoplasmic reticulum kinase; IRE1α, inositol-requiring enzyme-1; GCN2, general control nonderepressible 2; EIF2S1, eukaryotic translation initiation factor 2 subunit alpha; UPR, unfolded protein response; SEM, standard error of mean.
3.2 HMMR was associated with ER stress-driven HCC progression and discrepancy of HMMR expression at the mRNA and protein levels
To screen candidate genes involved in hepatitis-fibrosis-HCC progression, we performed RNA-seq on liver tissues from male HBV-tg mice and matched control mice aged 4, 10, and 16 months. The volcano plots of DEGs indicated that 267, 1,262, and 1,522 genes were significantly upregulated, while 100, 330, and 422 genes were significantly downregulated at 4, 10, and 16 months in the HBV-tg mice compared to the control mice, respectively (Supplementary Figure S3A). The total data set was obtained by merging 2,976 DEGs, which were further divided into 26 trend profiles by STEM analysis (Supplementary Figure S3B, Supplementary Figure S4). Among the profiles, profile 25, including 181 genes, displayed a continuously increasing trend (Supplementary Figure S4). The PPI network of the 181 continuously upregulated proteins in profile 25 was analyzed by the STRING database (Supplementary Figure S5). To identify the core protein in the network, we used the CytoHubba plugin to analyze the centrality of nodes. Based on 6 different centrality parameters, we selected HMMR with a high consistency score as the crucial candidate protein (Supplementary Table S4). Transcriptome sequencing of the mouse model showed that the mRNA levels of Hmmr continued to rise throughout the course of inflammation-carcinogenesis progression (Figure 2A). Therefore, we predicted Hmmr as an important candidate in ER stress-driven HCC progression.
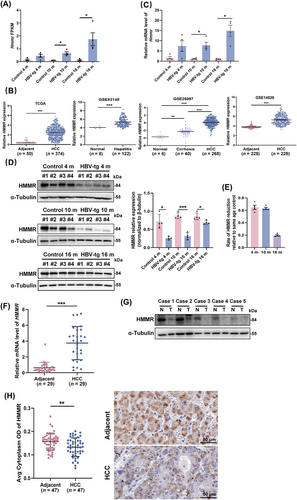
HMMR was associated with ER stress-driven HCC progression and discrepancies in HMMR expression at the mRNA and protein levels. (A) Relative expression of HMMR in transcriptomic data. (B) The mRNA expression of HMMR in HCC patients based on TCGA and GEO databases. (C) qRT-PCR analysis of HMMR mRNA expression in HBV-tg mice and control mice of the same age. (D) Protein level of HMMR in the liver tissue of the HBV-tg mice and semiquantitative analysis by Western blotting assay. (E) Rate of HMMR protein level reduction relative to the age-matched control group (n = 4). (F) qRT-PCR analysis of HMMR mRNA expression in the liver tissue of HCC patients (n = 29). (G) Protein level of HMMR in the liver tissue of HCC patients by Western blotting assay (n = 5). (H) IHC staining detected the expression of HMMR in the tumor and paired adjacent tissue of HCC patients (n = 47). Representative staining is shown. Two-tailed Student's t test or Welch's t test was used to test the significance of differences between two groups; data are represented as the mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Abbreviations: HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HMMR, hyaluronan-mediated motility receptor; SEM, standard error of mean.
To explore the role of HMMR in HCC, we analyzed its mRNA expression in hepatitis-fibrosis-HCC progression using the TCGA database and the GSE83148, GSE25097, and GSE14520 data sets. The mRNA level of HMMR continued to rise significantly throughout the course of progression (Figure 2B). We then validated the expression of HMMR in the liver of HBV-tg mice, and the qRT-PCR results showed that the mRNA expression of Hmmr was consistent with the increasing trend (Figure 2C). Surprisingly, the protein expression of HMMR was significantly reduced in the liver tissues of the HBV-tg mice compared to the age-matched control mice (Figure 2D). We also observed an interesting phenomenon in which the protein expression of HMMR in HBV-tg mice was reduced by 64.8% and 63.1% at 4 months and 10 months compared with that in the control mice of the same age, while 19.6% at 16 months (Figure 2E). Tumor tissue from patients with HCC yielded similar results that the mRNA expression of HMMR in liver tissues from HCC patients was significantly increased (Figure 2F), while the expression of HMMR protein was decreased in hepatic tumors compared with adjacent normal tissues by Western blotting analysis (Figure 2G). We also performed IHC to investigate the protein expression of HMMR in HCC tissues and adjacent normal tissues of 47 HBV-associated HCC patients, and the results were consistent with Western blotting analysis (Figure 2H). These results suggest that there may be a fine regulatory mechanism involved in HMMR expression under ER stress.
3.3 ER stress directly regulated the expression of HMMR via CHOP
In vitro induction using TM, a classical ER stress inducer, resulted in a time-dependent decrease in the protein expression of HMMR and a time-dependent increase in the mRNA expression of HMMR (Figure 3A-B), suggesting that ER stress may be the contributor regulating HMMR expression. Treatment with TUDCA, an ER stress repressor, blocked TM-induced HMMR mRNA elevation (Figure 3C), indicating that ER stress may account for the increase in HMMR transcription. Considering transcription factors that play an important transcriptional role in the UPR pathway, including XBP1s, ATF4, ATF6, and CHOP, we then tested the effect of these transcription factors on the transcriptional expression of HMMR. The results showed that CHOP specifically activated HMMR transcription (Figure 3D) and promoted HMMR expression in a dose-dependent manner (Figure 3E). We then investigated the binding of CHOP and HMMR promoter regions by luciferase assays. As shown in Figure 3F, CHOP-dependent HMMR promoter activation was significantly attenuated by the truncation of HMMR promoter. Silico prediction revealed several CHOP binding sites on the truncated region of the HMMR promoter, and the site with the highest score (-1297/-1282) was mutated (Figure 3G). The mutation blunted the transcriptional activation of CHOP on HMMR (Figure 3G-H). To show direct binding of CHOP to the predicted binding site within the HMMR promoter region, ChIP assay was performed. The result showed that CHOP could directly bind HMMR promoter containing domain between -1297/-1282, which was enhanced by TM stimulation (Figure 3I). Thus, these data suggest that CHOP is the transcription factor of ER stress that directly regulates HMMR expression.
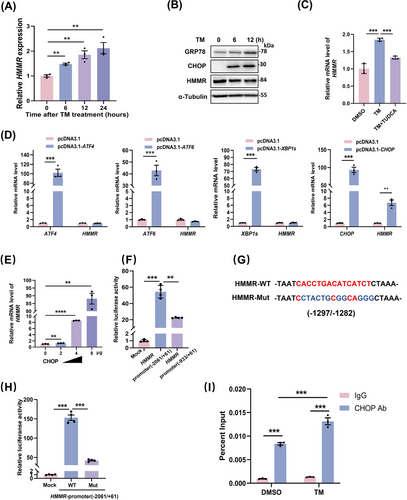
ER stress directly regulated the expression of HMMR by CHOP. (A) MHCC-97H cells were induced with TM (5 μg/mL) for the indicated times, and the mRNA expression of HMMR was detected by qRT-PCR. (B) MHCC-97H cells were induced with TM for the indicated times, and the protein levels of GRP78, CHOP, and HMMR were measured by Western blotting. (C) HLLCM3 cells treated with TUDCA (100 μmol/L) in the presence of TM (5 μg/mL) for 8 hours. HMMR expression was determined by qRT-PCR (n = 3). (D) MHCC-97H cells were transfected with ATF4, XBP1s, ATF6, or CHOP, and the expression of these transcription factors and HMMR was determined by qRT-PCR (n = 3). (E) HMMR expression in HCCLM3 cells was regulated by CHOP in a dose-dependent manner. (F) Luciferase activity of the full-length HMMR promoter reporter (-2061/+61) or HMMR promoter-truncated reporter (-933/+61) after cotransfection with CHOP in HEK293T cells (n = 4). (G) The predicted binding sites of CHOP in the HMMR promoter by JASPAR. (H) Luciferase activity of the full-length HMMR reporter or its mutant in binding sites after cotransfection with CHOP in HEK293T cells (n = 4). (I) ChIP analysis of CHOP binding at the HMMR promoter in MHCC-97H cells treated with or without TM (n = 3). Two-tailed Student's t test or Welch's t test was used to test the significance of differences between two groups; data are represented as the mean ± SEM. **, P < 0.01; ***, P < 0.001.
Abbreviations: Ab, antibody; ATF4, activating transcription factor 4; ATF6, activating transcription factor 6; ChIP, chromatin immunoprecipitation; CHOP, c/EBP homologous protein; DMSO, dimethyl sulfoxide; GRP78, glucose-regulated protein78; HMMR, hyaluronan-mediated motility receptor; MUT, mutant; TM, tunicamycin; TUDCA, tauroursodeoxycholate; WT, wild type; XBP1, X-box binding protein 1; SEM, standard error of mean.
3.4 HMMR was ubiquitinated by the E3 ubiquitin ligase TRIM29 under ER stress
We found that the mRNA level of HMMR was elevated under ER stress, and unexpectedly, the protein expression of HMMR was decreased under ER stress, which is consistent with the expression pattern of HMMR in HBV-tg mice liver compared with that in age-matched control group, suggesting that the expression of HMMR was subjected to post-translational regulation under ER stress. In the presence of CHX, HMMR degradation was blocked by the proteasome inhibitor MG132 but not by the lysosome inhibitor CQ (Figure 4A). MG132 increased HMMR protein expression under the condition of TM treatment (Figure 4B), suggesting that HMMR protein translation continued under ER stress. TM treatment obviously increased the ubiquitination level of HMMR (Figure 4C), suggesting that ER stress promoted ubiquitin-proteasome-mediated degradation of HMMR. To identify the E3 ubiquitin ligase that may ubiquitinate and degrade HMMR, we collected proteins that interacting with HMMR by Co-IP (Supplementary Figure S6A). Shotgun mass spectrometric assay identified several E3 ubiquitin ligases, in which RBX1 and SKP1 were present in both TM + MG132 and MG132 groups, while TRIM29 was present only in TM + MG132 group (Supplementary Table S5). Co-IP analysis validated that TRIM29 indeed interacted with HMMR (Figure 4D, Supplementary Figure S6B). Overexpression of TRIM29 enhanced ubiquitination of HMMR (Figure 4E, Supplementary Figure S6C). Immunofluorescence revealed a co-localization relationship between TRIM29 and HMMR, which can be enhanced with TM stimulation (Figure 4F, Supplementary Figure S6D). Therefore, TRIM29 was thought to be the E3 ubiquitin ligase that degrades HMMR.
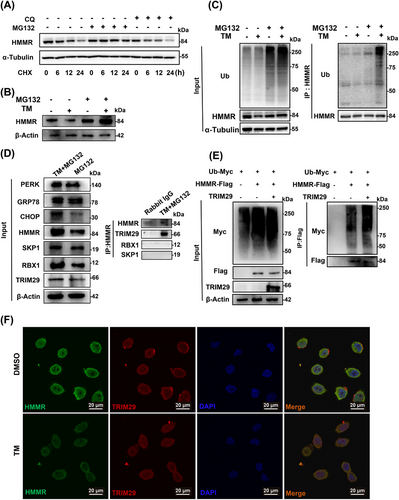
HMMR was ubiquitinated by the E3 ubiquitin ligase TRIM29 under ER stress. (A) MHCC-97H cells were treated with CQ or MG132 in the presence of CHX, and the protein level of HMMR was measured at the indicated times by Western blotting. (B) MHCC-97H cells were stimulated with TM (5 μg/mL) in the presence of MG132 (10 μmol/L) for 8 hours, and the protein level of HMMR was measured by Western blotting. (C) MHCC-97H cells were stimulated with MG132 and TM for 12 hours, and the ubiquitination of endogenous HMMR was detected by IP. (D) MHCC-97H cells were stimulated with TM in the presence of MG132 for 8 hours, and the interaction of HMMR and TRIM29 was detected by Co-IP and Western blotting. (E) MHCC-97H cells were cotransfected with ubiquitin-Myc, HMMR-Flag, and TRIM29 for 48 hours and then treated with MG132 for 6 hours. The protein lysates were immunoprecipitated to detect ubiquitination of HMMR. (F) Colocalization of HMMR and TRIM29 in HCCLM3 cells treated with TM by immunofluorescence assay.
Abbreviations: CHOP, c/EBP homologous protein; CHX, cycloheximide; CQ, chloroquine; DAPI, 4’,6-diamidino-2-phenylindole; DMSO, dimethyl sulfoxide; GRP78, glucose-regulated protein78; HMMR, hyaluronan-mediated motility receptor; PERK, protein kinase R-like endoplasmic reticulum kinase; Ub, ubiquitin; RBX1, ring-box 1; SKP1, S-phase kinase associated protein 1; TRIM29, tripartite motif containing 29; IP, immunoprecipitation; co-IP, coimmunoprecipitation.
3.5 HMMR was degraded by TRIM29 under ER stress
To further clarify the correlation between HMMR and TRIM29, we found that TRIM29 knockdown increased HMMR expression (Figure 5A), while TRIM29 overexpression decreased HMMR expression (Figure 5B). The expression of TRIM29 was knocked down with siTRIM29, and silencing TRIM29 extended the half-life of endogenous HMMR in the presence of CHX (Figure 5C); whereas overexpression of TRIM29 accelerated HMMR degradation and shortened the half-life of HMMR (Figure 5D). As expected, TRIM29 knockdown significantly rescued the protein expression of HMMR under ER stress induced by TM (Figure 5E).
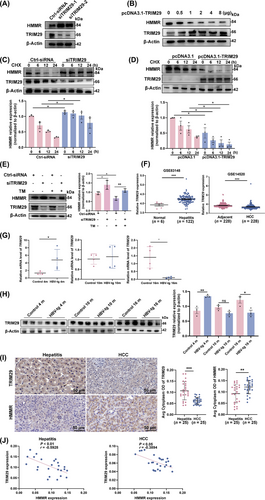
HMMR was degraded by TRIM29 under ER stress. (A) HCCLM3 cells were transfected with TRIM29-siRNAs for 48 hours, and the protein level of HMMR was measured by Western blotting. (B) HCCLM3 cells were transfected with pcDNA3.1-TRIM29 for 48 hours, and the protein level of HMMR was measured by Western blotting. (C) Control siRNA or TRIM29 siRNA was transfected into HCCLM3 cells for 36 hours, followed by incubation with CHX (100 μg/mL), and protein lysates were collected at the indicated times for western blot analysis. (D) pcDNA3.1 or pcDNA3.1-TRIM29 was transfected into HCCLM3 cells for 36 hours and then incubated with CHX (100 μg/mL), and protein lysates were collected at the indicated times for Western blotting analysis. (E) HCCLM3 cells were transfected with TRIM29-siRNA for 36 hours and then treated with TM, and the protein level of HMMR was measured by Western blotting. (F) The mRNA expression of TRIM29 in patients with liver hepatitis and HCC based on the GEO database. (G-H) qRT-PCR (G, n = 4) and Western blotting analysis (H, n = 3) of TRIM29 expression in the liver tissue of HBV-tg mice vs. control mice of the same age. (I) IHC staining detected the expression of HMMR and TRIM29 in human liver hepatitis tissues (n = 25) and HCC tissues (n = 25). Representative staining is shown. (J) Spearman correlation analysis between HMMR and TRIM29 based on the protein expression detected by IHC shown in (I). Western blot scanning densitometry for three independent experiments (C-E) or three biological repeats (H). Two-tailed Student's t test or Welch's t test was used to test the significance of differences between two groups; data are represented as the mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
Abbreviations: CHX, cycloheximide; GEO, gene expression omnibus; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HMMR, hyaluronan-mediated motility receptor; IHC, immunohistochemistry; SEM, standard error of mean; TRIM29, tripartite motif containing 29; TM, tunicamycin.
We then observed that the mRNA expression of TRIM29 was increased in hepatitis patients but decreased in HCC patients when compared with normal and adjacent tissues respectively using the GEO database (Figure 5F). Therefore, we analyzed the expression of TRIM29 in the liver tissues of HBV-tg mice by qRT-PCR and Western blotting. The results established that TRIM29 expression was higher in the liver of the HBV-tg hepatitis group than that of the same age control, while TRIM29 expression was lower in the liver of the HBV-tg HCC group than that of the same age control (Figure 5G-H). This may explain that the decrease rate of HMMR protein level in 16-month-old HBV-tg mice was lower than that in 4-month-old HBV-tg mice (Figure 2D-E). To further study, we examined the expression of HMMR and TRIM29 in human liver tissues of patients with HBV-related hepatitis and HCC. The results showed that compared with hepatitis tissues, the expression of HMMR was significantly higher in HCC tissues, while the expression of TRIM29 was significantly lower in HCC tissues (Figure 5I). Spearman correlation analysis revealed that in both hepatitis and HCC tissues, HMMR expression was negatively correlated with TRIM29 expression (Figure 5J), indicating a negative regulatory between TRIM29 and HMMR. These results demonstrated that HMMR was degraded by TRIM29 under ER stress.
3.6 HMMR alleviated ER stress by regulating autophagic flux and lysosomal activity
To further elucidate the role of HMMR during ER stress, we performed HMMR loss and gain of function experiments. Unexpectedly, we found that HMMR could regulate ER stress. Overexpression of HMMR inhibited TM-induced ER stress, while HMMR interference exacerbated TM-induced ER stress (Figure 6A-B). The expression of ER stress markers p-PERK/PERK and GRP78 were both significantly altered. To understand the mechanism of this phenomenon, we performed transcriptome sequencing on MHCC-97H cells that were stably transfected with HMMR. GSEA revealed that HMMR overexpression was involved in the regulation of autophagy (Supplementary Figure S7A). Thereafter, we detected 2 markers of autophagy, namely, LC3-II (reflecting the number of autophagosomes) and P62 (substrate of autophagy). Our results indicated that TM significantly increased the expression of LC3-II in a time-dependent manner, which was weakened by overexpression of HMMR, and the accumulation of P62 was significantly reduced by HMMR (Figure 6C). The TM-induced LC3-II increase was significantly enhanced after transient interference with HMMR, and P62 accumulation was significantly increased (Figure 6D). To further clarify whether HMMR regulates autophagosome formation or autophagolysosome activity, we used the lysosome inhibitors CQ and Baf A1 under ER stress induction. After inhibition of lysosomes, the regulatory effects of HMMR on LC3-II and P62 disappeared (Figure 6E-F), which indicated that HMMR may regulate autophagy by promoting lysosomal degradation.
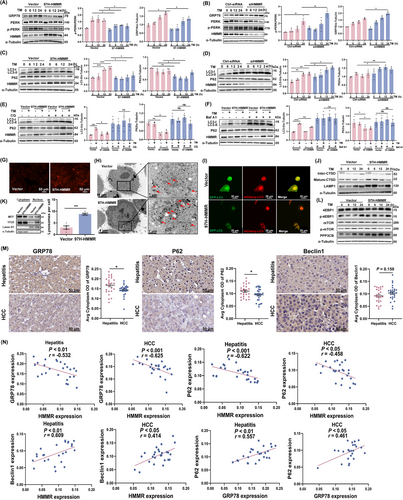
HMMR alleviated ER stress by regulating autophagic flux and lysosomal activity. (A-B) MHCC-97H cells were overexpressed with HMMR (A) or transfected with HMMR-siRNAs (B) and induced with TM. The protein levels of GRP78, PERK, p-PERK, and HMMR were measured at the indicated times by Western blotting. (C-D) MHCC-97H cells were overexpressed with HMMR (C) or transfected with HMMR-siRNAs (D) and then treated with TM. The protein levels of LC3-I/II, p62, and HMMR were measured at the indicated times by Western blotting. (E) MHCC-97H cells were overexpressed with HMMR and induced with TM or CQ for 12 hours, and the protein levels of LC3-I/II, p62, and HMMR were measured by Western blotting. (F) MHCC-97H cells were overexpressed with HMMR and induced with TM or Baf A1 for 12 hours, and the protein levels of LC3-I/II, p62, and HMMR were measured by Western blotting. (G) HMMR overexpressing MHCC-97H cells were stained with LysoTracker Red and observed under fluorescence microscope. (H) Transmission electron scanning showed lysosomes in MHCC-97H cells overexpressing HMMR. Lysosomes are pointed by the arrows. Semiquantitative analysis of the lysosome counts is shown in the histogram to the left of transmission electron scanning image. (I) HMMR overexpressing MHCC-97H cells were transfected with the dual-labelled mCherry-GFP-LC3 plasmid for 24 hours and observed under confocal laser scanning microscope. (J) MHCC-97H cells were overexpressed with HMMR and induced with TM, and the protein levels of CTSD and LAMP1 were measured at the indicated times by Western blotting. (K) Nuclear and cytoplasmic proteins were extracted from MHCC-97H cells overexpressing HMMR, and the protein levels of TFEB and MITF were measured by Western blotting. (L) MHCC-97H cells were overexpressed with HMMR and induced with TM, and the protein levels of 4EBP1, p-4EBP1, mTOR, p-mTOR, and PPP3CB were measured at the indicated times by Western blotting. (M) IHC staining detected the expression of GRP78, P62, and Beclin1 in human liver hepatitis tissues (n = 25) and HCC tissues (n = 25). Representative staining is shown. (N) Spearman correlation analysis between HMMR and GRP78, HMMR and P62, HMMR and Beclin1, and GRP78 and P62 based on the protein expression detected by IHC shown in (M) and Figure 5I. Western blot scanning densitometry for three independent experiments (A-F). Two-tailed Student's t test or Welch's t test was used to test the significance of differences between two groups; data are represented as the mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Abbreviations: 4EBP, eukaryotic translation initiation factor 4E-binding protein 1; Baf A1, bafilomycin A1; CQ, chloroquine; CTSD, cathepsin D; GRP78, glucose-regulated protein78; GFP, green fluorescent protein; HMMR, hyaluronan-mediated motility receptor; HCC, hepatocellular carcinoma; LAMP1, lysosome-associated membrane protein 1; LC3, microtubule associated protein 1 light chain 3; MITF, microphthalmia-associated transcription factor; mTOR, mechanistic target of rapamycin kinase; PERK, protein kinase R-like endoplasmic reticulum kinase; P62/SQSTM1, sequestosome 1; PPP3CB, protein phosphatase 3 catalytic subunit beta; SEM, standard error of mean; TM, tunicamycin; TFEB, transcription factor EB.
To further clarify the mechanism by which HMMR regulates lysosomal activity, we performed lysosome staining and electron microscopy and found that the number of lysosomes was significantly increased after HMMR overexpression (Figure 6G-H). We evaluated the autophagy flux in HMMR overexpressing cells. The results showed that HMMR overexpressing cells exhibited abundant red puncta (representing autolysosome) formation compared with control group (Figure 6I), which validated our conclusion that HMMR activated autophagy by promoting the formation of autophagic lysosomes. Moreover, the protein expression of mature cathepsin D (CTSD) and lysosomal associated membrane protein 1 (LAMP1), which reflect lysosomal activity, was increased (Figure 6J). We found that HMMR overexpression promoted the nuclear translocation of lysosome-related transcription factors, including transcription factor EB (TFEB) and melanocyte inducing transcription factor (MITF) (Figure 6K). TFEB nuclear translocation depends on its phosphorylation, which is regulated by two pathways. The mTOR pathway promotes the phosphorylation of TFEB to inhibit the nuclear translocation of TFEB, while protein phosphatase 3 catalytic subunit beta (PPP3CB) dephosphorylates TFEB and induces TFEB nuclear translocation [34, 35]. GSEA suggested that HMMR overexpression induced phosphatase complex activity (Supplementary Figure S7B). Western blotting assay indicated that PPP3CB expression was obviously increased after HMMR overexpression, while there was no change in the mTOR signaling pathway (Figure 6L), suggesting that HMMR overexpression promoted TFEB nuclear translocation by upregulating PPP3CB expression, thus promoting lysosomal activity.
We next conducted IHC to detect the expression of HMMR, ER stress marker GRP78, and autophagy markers P62 and Beclin1 in human liver tissues of patients with HBV-related hepatitis and HCC. The results showed that compared with hepatitis tissues, the expression of HMMR was significantly higher in HCC tissues, but the expression of GRP78 and P62 was significantly lower, while the expression of Beclin1 showed an increasing trend (Figure 5I, Figure 6M). The expression pattern of HMMR in human liver hepatitis and HCC tissues was similar with that in HBV-tg mouse model (Figure 5, 4 m and 16 m of Figure 2D). We then analyzed the correlation between the expression of these molecules. Spearman correlation analysis revealed that in both hepatitis and HCC tissues, HMMR expression was negatively correlated with the expression GRP78 and autophagic substrate P62 respectively, while positively correlated with Beclin1 expression (Figure 6N). In addition, the expression of GRP78 and P62 was positively correlated, suggesting a negative regulation of ER stress intensity and autophagic activity. Taken together, these results suggested that HMMR alleviated ER stress by regulating autophagy.
In order to demonstrate the role of HMMR by regulating autophagy and ER stress in HCC cell proliferation, we performed CCK8 assay. As shown in Supplementary Figure S8, overexpression of HMMR promoted cell proliferation, which could be retarded with autophagy inhibitor CQ or ER stress inducer TM, even further being more arrested by both interventions.
4 DISCUSSION
Prolonged and attenuated ER stress may stimulate HBV-related inflammatory-cancer transformation. In the present study, we proposed that HMMR may alleviate ER stress by promoting autophagolysosomal activity during ER stress-driven HCC progression. Several novel aspects were identified in this work. First, transcription of HMMR persistently increased during ER stress-driven HCC progression, which is directly regulated by CHOP, a downstream transcription factor of UPR pathway. Second, under ER stress, the altered expression of HMMR protein levels is dynamically controlled by E3 ligase TRIM29, resulting in ubiquitinated degradation. Finally, HMMR promotes autophagolysosomal activity by inducing nuclear translocation of lysosome-related transcription factor, TFEB, leading to attenuation of ER stress. Thus, our studies indicate that the dynamic degradation of HMMR under ER stress leads to sustained and attenuated ER stress, which may provide a favorable environment for HBV-related cancer transformation (Figure 7).
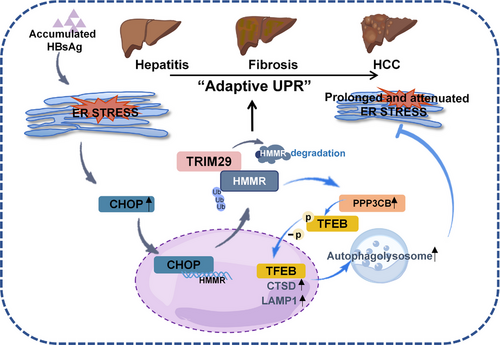
A proposed model for the role of HMMR in interaction of ER stress and autophagy during HCC progression.
Abbreviations: CHOP, c/EBP homologous protein; CTSD, cathepsin D; ER, endoplasmic reticulum; HBsAg, hepatitis B surface antigen; HCC, hepatocellular carcinoma; HMMR, hyaluronan-mediated motility receptor; LAMP1, lysosome-associated membrane protein 1; PPP3CB, protein phosphatase 3 catalytic subunit beta; TFEB, transcription factor EB; TRIM29, tripartite motif containing 29; Ub, ubiquitin; UPR, unfolded protein response.
It is an objective fact that gender differences in the incidence of HCC in epidemiology [2, 36, 37]. There have been several studies on the mechanisms of the role that sex hormones played in HCC [38, 39]. Interestingly, in our study of the natural course of hepatitis-fibrosis-HCC progression using an HBV-tg mouse model, we found remarkably differences in the course of the disease between the genders, that male mice developed pathological changes earlier and more severely than female mice (Supplementary Figure S1, Supplementary Figure S2, Supplementary Table S3). This also validated the used animal model displaying the characteristics of clinical HCC development.
The UPR is an important mechanism required for cancer cells to adapt to the transformed state and maintain malignancy [40]. The accumulation of unfolded and misfolded proteins induces a continuous accumulation of proteins in the ER, triggering a “chronic ER stress response”. Cells gradually adapt to the stressful state, and the adaptive UPR promotes the completion of malignant transformation of cells [41]. In this study, we clarified the sustained activation of ER stress induced by overexpression of LHB throughout the hepatitis-fibrosis-cancer process in an HBV-tg mouse model. This provided a clear basis for the direct mechanism of HBV action on malignant transformation of hepatocytes throughout inflammatory cancer progression. Moreover, ER stress is more severe in the early stages of the liver disease process than in the later stages (Figure 1F), which is the key for cells to be able to adapt to the UPR rather than to undergo cell death. How do cells regulate this process so that the stress state is instead hijacked as a tool for malignant transformation? We used transcriptomic techniques and identified HMMR, which may play an essential role in the whole process. Interestingly, we discovered that HMMR mRNA was consistently elevated over the course of the disease; however, HMMR protein expression was not. We believe that the dynamic regulation of molecular expression by ER stress is a crucial factor in accomplishing its malignant transformation.
The tripartite motif-containing protein (TRIM) family, a family of proteins with a relatively conserved structure that contain RING-finger domain proteins, plays an important role in tumorigenesis [42]. TRIM29 is associated with the β-catenin pathway in HCC [43]. However, the role of the E3 ubiquitin ligase TRIM29 has not been reported in HCC. The regulation of protein expression is undoubtedly a complex process. ER-associated degradation E3 ligases, including HRD1 [44], gp78 [45, 46], and RMA1 [46], have been reported to degrade tumor suppressor proteins. Our study clarified the degradation of HMMR by TRIM29, which is identified as a novel ERAD E3 ligase under ER stress (Figure 4D-F, Figure 5A-E). As there was a transition from high to low TRIM29 expression during the progression of inflammatory-cancer transformation (Figure 5F-I), the protein level of HMMR was associated with TRIM29 expression. The expression of TRIM29 and HMMR in human hepatitis and HCC tissues were detected and confirmed they were both negatively correlated (Figure 5I-J). We identified for the first time that this pair of molecular switches and the resulting feedback regulation of ER stress as well as autophagy may lead the UPR pathway to a procarcinogenic point of no return.
Autophagy plays a dual role in liver disease and HCC [47, 48] and controls protein quality by digesting excess protein and recycling faulty cellular components [49]. HBV might induce autophagosome formation as its replication site to facilitate the viral life cycle but block subsequent autophagic-lysosome fusion to evade autophagic degradation [50]. Intriguingly, our findings indicated that the TM-induced increase in LC3-II was attenuated and that the accumulation of P62 was significantly reduced after HMMR overexpression (Figure 6C), and HMMR activated lysosomal activity (Figure 6G-I). In addition, we further found that HMMR regulates lysosomal activity by modulating the nuclear translocation of TFEB. TFEB is the main transcription factor that regulates the expression of lysosomes and autophagy-related genes. Nuclear translocation of TFEB is associated with its dephosphorylation. Phosphorylation of TFEB is mediated by mTORC1 and retains TFEB in the cytoplasm [34]. In contrast, PPP3CB, a serine/threonine protein phosphatase, dephosphorylates TFEB and promotes the nuclear translocation of TFEB [51]. We demonstrate that HMMR promotes nuclear translocation of TFEB through the PPP3CB pathway, which activates lysosome-associated transcription factors and thus activates lysosomal activity. This finding clearly elucidates the mechanism by which HMMR regulates autophagy.
As autophagy could act as a degradation system for unfolded proteins accumulated in the ER in addition to ERAD [52], increased activity of autophagic lysosomes can reduce protein load and therefore alleviate the UPR [53]. The degradation of HMMR via the ERAD pathway can be alleviated by the relief of ER stress (Figure 2E). That is, our study identified a mechanism by which HMMR can stabilize itself through the dual regulation of ER stress and autophagy. We found new crosstalk between the UPR and autophagy in which HMMR alleviates ER stress precisely by activating lysosomal activity. This dual regulation of the extent of UPR activity allows the UPR pathway to persist at a proportionate level, thus allowing the cells to undergo malignant transformation.
Although our study provided a completely mechanisms of the inconsistency expression of HMMR in mRNA and protein level, as well as describing an interesting interaction of HMMR, ER stress, and autophagy during HCC progression, there were still some issues to be further investigated. (1) The ER stress that we observed in HBV-tg mice was caused by HBV LHB overexpression, and we also observed that LHB overexpression could induce ER stress (data not show), thus the specific regulatory mechanism of HMMR under LHB-induced ER stress in HCC development remains to be further studied. (2) We demonstrated that HMMR promoted lysosome activity via upregulation of PPP3CB, however its specific mechanisms have not been fully elucidated.
5 CONCLUSIONS
In summary, the present study revealed a previously unappreciated role of HMMR as a central node for transcriptional activation by CHOP and degradation by TRIM29 under ER stress, and as a feedback mechanism, HMMR alleviates ER stress by activating autophagic lysosomes. Oncogenic pathways involve strategies to manipulate ER stress and autophagy for their own benefit that use such a pair of molecular switches to regulate the intensity of the adaptive UPR. HMMR may be a novel therapeutic target in the context of strategies targeting the UPR pathway for HBV-associated HCC prevention.
DECLARATIONS
AUTHOR CONTRIBUTIONS
Huijie Bian and Zhi-Nan Chen conceived and designed the project designed the study and revised manuscript. Lin He and Hao Li performed most of the experiments and drafted the original manuscript. Lin He, Hao Li, and Can Li performed histological and animal experiments. Ze-Kun Liu, Hao Li, and Hao-Lin Wei provided bioinformatics analysis. Meng Lu, Ren-Yu Zhang, Man Liu, Dong Wu, Jie Shao, Zhe Wang, and Cong Zhang contributed technical/reagents materials. Huijie Bian, Ling-Min Kong, and Ding Wei reviewed and/or revised the manuscript. All authors approved the final manuscript.
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Ying Liu (Institute of Basic Medical Sciences Chinese Academy of Medical Sciences) for providing us ATF6 plasmid.
CONFLICT OF INTERESTS STATEMENT
The authors declare that they have no competing interests.
FUNDING INFORMATION
This study was supported by the National Natural Science Foundation of China (82130084) and Shaanxi Provincial Key R&D Program (2021SF-110).
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Ethical approval was obtained from the Ethics Committee of the Fourth Military Medical University (202003-216). Tissue samples were obtained with written informed consent from each patient. All animal studies were approved by the Animal Care and Use Committee of National Translational Science Center for Molecular Medicine (2020-NTSCMM-ID004).
CONSENT FOR PUBLICATION
Not applicable.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




