Cytoplasmic YAP1-mediated ESCRT-III assembly promotes autophagic cell death and is ubiquitinated by NEDD4L in breast cancer
These authors contributed equally: Yan Guo, Yuqing Cui, and Yangyang Li.
Abstract
Background
Nuclear Yes1-associated transcriptional regulator (YAP1) promotes tumor progression. However, the function of cytoplasmic YAP1 in breast cancer cells and its impact on the survival of breast cancer patients remain unclear. Our research aimed to explore the biological function of cytoplasmic YAP1 in breast cancer cells and the possibility of cytoplasmic YAP1 as a predictive marker of breast cancer survival.
Methods
We constructed cell mutant models, including NLS-YAP15SA (nuclear localized), YAP1S94A (incapable of binding to the TEA domain transcription factor family) and YAP1S127D (cytoplasmic localized), and used Cell Counting Kit-8 (CCK-8) assays, 5-ethynyl-2’-deoxyuridine (EdU) incorporation assays, and Western blotting (WB) analysis to detect cell proliferation and apoptosis. The specific mechanism of cytoplasmic YAP1-mediated endosomal sorting complexes required for transport III (ESCRT-III) assembly was studied by co-immunoprecipitation, immunofluorescence staining, and WB analysis. Epigallocatechin gallate (EGCG) was used to simulate YAP1 retention in the cytoplasm in in vitro and in vivo experiments to study the function of cytoplasmic YAP1. YAP1 binding to NEDD4-like E3 ubiquitin protein ligase (NEDD4L) was identified using mass spectrometry and was verified in vitro. Breast tissue microarrays were used to analyze the relationship between cytoplasmic YAP1 expression and the survival of breast cancer patients.
Results
YAP1 was mainly expressed in the cytoplasm in breast cancer cells. Cytoplasmic YAP1 promoted autophagic death of breast cancer cells. Cytoplasmic YAP1 bound to the ESCRT-III complex subunits charged multivesicular body protein 2B (CHMP2B) and vacuolar protein sorting 4 homolog B (VPS4B), promoting assembly of CHMP2B-VPS4B and activating autophagosome formation. EGCG retained YAP1 in the cytoplasm, promoting the assembly of CHMP2B-VPS4B to promote autophagic death of breast cancer cells. YAP1 bound to NEDD4L, and NEDD4L mediated ubiquitination and degradation of YAP1. Breast tissue microarrays revealed that high levels of cytoplasmic YAP1 were beneficial to the survival of breast cancer patients.
Conclusions
Cytoplasmic YAP1 mediated autophagic death of breast cancer cells by promoting assembly of the ESCRT-III complex; furthermore, we established a new breast cancer survival prediction model based on cytoplasmic YAP1 expression.
Abbreviations
-
- YAP1
-
- nuclear Yes1-associated transcriptional regulator
-
- CCK-8
-
- Cell Counting Kit-8
-
- EdU
-
- 5-Ethynyl-2’- deoxyuridine
-
- WB
-
- western blotting
-
- ESCRT
-
- endosomal sorting complexes required for transport
-
- co-IP
-
- co-immunoprecipitation
-
- IF
-
- immunofluorescence staining
-
- EGCG
-
- epigallocatechin gallate
-
- NEDD4L
-
- NEDD4-like E3 ubiquitin protein ligase
-
- CHMP2B
-
- charged multivesicular body protein 2B
-
- VPS4B
-
- vacuolar protein sorting 4 homolog B
-
- CTGF
-
- connective tissue growth factor
-
- CYR61
-
- cysteine rich angiogenic inducer 61
-
- ANKRD1
-
- ankyrin repeat domain 1
-
- TEAD
-
- TEA domain transcription factor
-
- AP
-
- autophagosome
-
- AL
-
- autolysosome
-
- HMGB1
-
- high mobility group box 1
-
- ROS
-
- reactive oxygen species
-
- PI3K
-
- phosphatidylinositol 3-kinase
-
- ATG4
-
- autophagy-related gene 4
-
- HIF-1α
-
- hypoxia-inducible factor
-
- mTOR
-
- mechanistic target of rapamycin kinase
-
- MAPK
-
- mitogen-activated protein kinase
-
- IHC
-
- immunohistochemistry
-
- SQSTM1/p62
-
- sequestosome 1
-
- MOB1A
-
- MOB kinase activator 1A
-
- p-MOB1A
-
- phosphorylated-MOB1A
-
- LC3
-
- microtubule associated protein 1 light chain 3
-
- IgG
-
- immunoglobulin G
-
- MST1
-
- macrophage stimulating 1
-
- PARP
-
- poly (ADP-ribose) polymerase 1
-
- Ub
-
- ubiquitin
-
- ATG7
-
- autophagy related 7
-
- CQ
-
- chloroquine
-
- CHX
-
- cycloheximide
-
- EBSS
-
- Earle's Balanced Salt Solution
-
- DMEM
-
- dulbecco's modified eagle medium
-
- siRNA
-
- small interfering RNA
-
- mRFP
-
- monomer red fluorescent protein
-
- GFP
-
- green fluorescent protein
-
- EGFP
-
- enhanced green fluorescent protein
-
- RIPA
-
- radio-immunoprecipitation assay
-
- PMSF
-
- phenylmethylsulfonyl fluoride
-
- SDS
-
- sodium dodecyl sulfate
-
- PAGE
-
- polyacrylamide gel electrophoresis
-
- PVDF
-
- polyvinylidene fluoride
-
- ECL
-
- enhanced chemiluminescence
-
- DAB
-
- 3,3′-diaminobenzidine
-
- OD450
-
- optical density at 450nm
-
- PBS
-
- phosphate-buffered saline
-
- qRT-PCR
-
- quantitative real-time PCR
-
- GAPDH
-
- glyceraldehyde-3-phosphate dehydrogenase
-
- PI
-
- propidium iodide
-
- PPI
-
- protein-protein interaction
-
- DAPI
-
- 4',6-diamidino-2-phenylindole
-
- TEM
-
- transmission electron microscopy
-
- EDTA
-
- ethylene diamine tetraacetic acid
-
- TCGA
-
- The Cancer Genome Atlas
-
- HPA
-
- Human Protein Atlas
-
- NPASS
-
- Natural Product Activity & Species Source Database
-
- GEO
-
- Gene Ommnibus
-
- DEG
-
- differentially expressed gene
-
- WGCNA
-
- weighted gene co-expression analysis
-
- KEGG
-
- Kyoto Encyclopedia of Genes and Genomes
-
- ROC
-
- receiver operating characteristic
-
- AUC
-
- area under the curve
-
- ANOVA
-
- analysis of variance
-
- RFS
-
- relapse-free survival
-
- DFS
-
- disease-free survival
-
- OS
-
- overall survival
-
- ER+
-
- estrogen receptor-positive
-
- HER2+
-
- human epidermal growth factor receptor 2-positive
-
- ESR1
-
- estrogen receptor 1
-
- VGLL3
-
- vestigial like family member 3
-
- PAS
-
- phagophore assembly site
-
- PG
-
- elongation of the phagophore
-
- ULK1
-
- unc-51 like autophagy activating kinase 1
-
- PE
-
- phosphatidylethanolamine
-
- MIT
-
- microtubule interaction and transport
-
- β-TRCP
-
- beta-transducin repeat containing E3 ubiquitin protein ligase
-
- Not4
-
- CCR4-NOT core ubiquitin-protein ligase subunit
-
- USP10
-
- ubiquitin specific peptidase 10
-
- USP47
-
- ubiquitin specific peptidase 47
-
- OTUB2
-
- OUT deubiquitinase, ubiquitin aldehyde binding 2
-
- EIF3H
-
- eukaryotic translation initiation factor 3 subunit H
-
- HR
-
- hazard ratio
-
- CI
-
- confidence interval
-
- YAP1WT
-
- YAP1wild typ
-
- NLS-YAP15SA
-
- nuclear localization sequence-YAP15SA
-
- EtOH
-
- ethanol
-
- E2
-
- estrogen
-
- COPII
-
- the Coat Protein complex II
1 BACKGROUND
Breast cancer ranks first in female tumor incidence worldwide, and the mortality is second only to lung cancer [1, 2]. Over the years, many studies have been conducted to find suitable mechanisms and targets for breast cancer treatments to improve the survival rate of breast cancer patients. Yes1-associated transcriptional regulator (YAP1) is a classic molecule downstream of the Hippo pathway. Previous studies largely focused on transcriptional coactivation of nuclear YAP1, which promoted transcription of the downstream pro-proliferation genes connective tissue growth factor (CTGF), cysteine rich angiogenic inducer 61 (CYR61), and ankyrin repeat domain 1 (ANKRD1) by binding to members of the TEA domain transcription factor (TEAD) family, and these pro-proliferation genes mediated malignant phenotypes and tumor proliferation [3, 4].
Despite the results discussed above, different views exist regarding the role of YAP1 in tumors. Some studies have found that the expression of YAP1 in the cytoplasm inhibited the proliferation of colorectal cancer [5-7]. In the cytoplasm, YAP1 interacted with the Wnt/β-catenin pathway, inhibiting both the translocation of β-catenin into the nucleus and subsequent proliferation of tumor cells [6, 8]. In the nucleus, YAP1 bound to the tumor suppressor P73, inhibiting ubiquitination and degradation of P73 and thus stabilizing it; this promoted the transcription of P73-induced apoptotic genes and tumor cell apoptosis [9-12]. A recently published pan-cancer analysis revealed that tumors can be classified based on YAP1 silencing; in the YAP1-silenced tumors, YAP1 acted as a tumor suppressor, while in the YAP1-expressed tumors, YAP1 acted as an oncogene [13]. Therefore, the effects of YAP1 on tumor cell fate remain to be clarified.
Autophagy is an evolutionarily conserved catabolic degradation process in cells, a form of programmed cell death that is distinct from apoptosis. Continuous activation of autophagy can lead to degradation of necessary cellular components and promotion of cell death [14]. The process of autophagy involves several main stages, including activation of autophagy, formation of the autophagosome (AP), and fusion of the AP and lysosome to form the autolysosome (AL) [15]. Overexpression of YAP1 promoted transcription and cytoplasmic localization of the positive autophagy regulator high mobility group box 1 (HMGB1) [16]; YAP1 overexpression also promoted autophagy by activating reactive oxygen species (ROS) [16, 17]. HMGB1 and ROS both induced autophagy indirectly by activating signaling pathways. In cytoplasm, the Beclin-1/phosphatidylinositol 3-kinase (PI3K) III complex was activated by HMGB1 to promote autophagy [18, 19], and ROS acted as signaling molecules in the regulation of autophagy by targeting autophagy genes (e.g., autophagy-related gene 4 [ATG4]), transcription factors (e.g., hypoxia-inducible factor-1α [HIF-1α]), and signal transduction systems (e.g., mechanistic target of rapamycin kinase [mTOR] and mitogen-activated protein kinases [MAPKs]) [20]. However, whether there is a more direct mechanism by which YAP1 activates autophagy remains unclear. Each step of autophagy is related, to varying degrees, to dynamic processes such as vesicle separation and transport. Studies have found that the endosomal sorting complexes required for transport (ESCRTs), which are involved in membrane shearing and sealing, promoted the sealing of AP membranes, thereby promoting the formation of APs [21-23]. The function of ESCRT primarily requires three sub-complexes: ESCRT-I, ESCRT-II, and ESCRT-III. When the AP membrane was closed, ESCRT-III was recruited to the double bowl-shaped membrane; vacuolar protein sorting 4 homolog B (VPS4B) later bound to ESCRT-III, then remained bounding to the complex near the open bowl-shaped bilayer membrane for 1-2 min to seal it, creating a complete AP [24].
The present study explored the effect of cytoplasmic YAP1 on the autophagic death of breast cells and its value on the survival of breast cancer patients. We focused on the subcellular localization of YAP1 to precisely study the function of cytoplasmic YAP1 in breast cancer. By transfecting mutant NLS-YAP15SA (nuclear localized), mutant YAP1S94A (incapable of binding to TEAD transcription factor family) and mutant YAP1S127D (cytoplasmic localized), cell proliferation and apoptosis were detected. The specific mechanism of cytoplasmic YAP1-mediated ESCRT-III assembly was studied by co-immunoprecipitation (co-IP), immunofluorescence (IF) staining and Western blotting (WB) analysis. The clinical specimens from breast cancer patients were used to analyze the relationship between cytoplasmic YAP1 expression and the survival of breast cancer patients.
2 MATERIALS AND METHODS
2.1 Antibodies and reagents
Antibodies for YAP1 (13584-1-AP, 66900-1-Ig; 1:1000 for WB, 1:200 for immunohistochemistry [IHC], 1:100 for IF), NEDD4 like E3 ubiquitin protein ligase (NEDD4L, 13690-1-AP, 1:1000 for WB, 1:400 for IHC), sequestosome 1 (SQSTM1/p62, 18420-1-AP, 1:1000 for WB, 1:50 for IHC) and CYR61 (26689-1-AP, 1:1000 for WB, 1:50 for IHC) were purchased from Proteintech Group (Rosemont, IL, USA). VPS4B antibodies (sc-377162, 1:1000 for WB, 1:50 for IF) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). MOB kinase activator 1A (MOB1A, AF0730, 1:1000 for WB) and phosphorylated MOB1A antibodies (p-MOB1A, AF4481, 1:500 for WB) were purchased from Affinity Biosciences (Cincinnati, OH, USA). Charged multivesicular body protein 2B (CHMP2B, 76173, 1:200 for WB), microtubule-associated protein 1 light chain 3 A/B (LC3A/B, 4108, 12741, 1:1000 for WB, 1:100 for IHC), p-YAP1 (13008, 1:500 for WB), macrophage stimulating 1 (MST1, 3682, 1:1000 for WB), p-MST1 (49332, 1:500 for WB), and normal rabbit immunoglobulin G (IgG, 2729, 1:400 for co-IP) were purchased from Cell Signaling Technology (Boston, MA, USA). CHMP2B antibodies (ab157208, 1:500 for WB, 1:50 for IF) were purchased from Abcam (Cambridge, England). CTGF (WL02602, 1:500 for WB), Caspase-3 (WL02117, 1:500 for WB), poly(ADP-ribose) polymerase 1 (PARP1, WL01932, 1:500 for WB), and ubiquitin (Ub) antibodies (WL01368, 1:500 for WB) were purchased from Wanleibio (Shenyang, Liaoning, China). Autophagy-related 7 (ATG7) antibodies (T55658, 1:1000 for WB) were purchased from Abmart Incorporate (Shanghai, China). CTGF antibodies (PB0570, 1:50 for IHC) were purchased from Boster (Beijing, China). β-actin (TA-09, 1:1000 for WB), goat anti-rabbit IgG (ZB-2301, 1:5000 for WB), and goat anti-mouse IgG (ZB-2305, 1:5000 for WB) were purchased from ZSGB-Bio (Beijing, China). Chloroquine (CQ, HY-17589A), cycloheximide (CHX, HY-12320), MG132 (HY-13259), and XMU-MP-1 (HY-100526) were purchased from MedChemExpress (Shanghai, China). Epigallocatechin gallate (EGCG, E4143) was purchased from Sigma-Aldrich (St. Louis, MO, USA). Earle's Balanced Salt Solution (EBSS, PB180337) was purchased from Procell Life Science & Technology Co., Ltd. (Wuhan, Hubei, China).
2.2 Cell culture
The normal mammary epithelial cell line MCF-10A and breast cancer cell lines (T47D, MCF7, SKBR3, MDA-MB-453, UACC-812, MDA-MB-231, BT-549, and MDA-MB-468) were obtained from the Heilongjiang Cancer Institute (Harbin, Heilongjiang, China). All cell lines were authenticated using the short tandem repeat method by the Procell Life Science & Technology Company in 2016. The MCF-10A cell line was cultured in complete medium (CM-0525, Procell). T47D, MCF7, and UACC-812 cell lines were cultured in Dulbecco's Modified Eagle Medium (DMEM) (Gibco, Carlsbad, CA, USA). SKBR3, MDA-MB-453, and BT-549 cell lines were cultured in RPMI-1640 (Gibco). MDA-MB-231 and MDA-MB-468 cell lines were cultured in Leibovitz's L15 medium (PM151010, Procell) supplemented with 10% fetal bovine serum (FSP500, ExCell Bio, Suzhou, Jiangsu, China) and 1% penicillin-streptomycin (PYG0016, Boster). The MDA-MB-231 and MDA-MB-468 cell lines were cultured at 37°C in a sterile incubator containing no CO2; all other cell lines were cultured at 37°C in a sterile incubator containing 5% CO 2.
2.3 Plasmids, lentivirus and transfection
VPS4B (G116254) overexpression plasmids were purchased from YouBio (Changsha, Hunan, China). YAP1 (HG17690-UT), CHMP2B (HG14596-UT), and NEDD4L (HG17684-UT) overexpression plasmids were purchased from Sino Biological Inc. (Beijing, China). NLS-YAP15SA, YAP1S94A, YAP1S127D, ΔYAP1, YAP1R87A, and YAP1F95A overexpression plasmids were constructed by and enhanced green fluorescent protein (EGFP) control plasmid were purchased from Genechem (Shanghai, China). Truncated mutants of YAP1 including N (1-159aa), WW (109-324aa), C (275-504aa) were constructed by Hanyin Biotechnology (Shanghai, China). Small interfering RNAs (siRNAs) were obtained from RiboBio (Guangzhou, Guangdong, China). Adenovirus of monomer red fluorescent protein (mRFP)-green fluorescent protein (GFP)-LC3 was purchased from Hanbio Biotechnology (Shanghai, China).
Cells were grown using the conditions described above. When cells had grown to 60%–70% density, the transfection reagent jetPRIME® (101000046, Polyplus-transfection SA, Strasbourg, France) was used to transiently transfect cells with overexpression plasmids or siRNAs following the manufacturer's instructions. mRFP-GFP-LC3 was transfected without any transfection reagents. Lentivirus-YAP1 was stably transfected according to manufacturer's instructions, and puromycin was used for stability screening. Experiments were carried out 24-72 h after transfection.
The sequences targeted by the siRNAs were as follows: siYAP1#1 (5'-GAGATGGAATGAACATAGA-3'), siYAP1#2 (5'-GCGTAGCCAGTTACCAACA-3'), siYAP1#3 (5'-CAGTGGCACCTATCACTCT-3'); siATG7#1 (5'-GAACGAGTATCGGCTGGAT-3'), siATG7#2 (5'-GATGTCGTCTTCCTATTGA-3'), siATG7#3 (5'-ACTCGAGTCTTTCAAGACT-3'); siNEDD4L#1 (5'-GGAGAATTATGTCCGTGAA-3'), siNEDD4L#2 (5'-GAACCCTCCTCAAGGTTGA-3'), siNEDD4L#3 (5'-GGCCGAACTTACTATGTCA-3').
2.4 Patient information and tissue specimens
All normal and breast cancer tissues were obtained from patients who underwent surgeries in 2007 at Harbin Medical University Cancer Hospital (Harbin, Heilongjiang, China). Six pairs of fresh breast cancer tissues and matched adjacent normal breast tissues were used to analyze the expression of YAP1 and NEDD4L by WB analysis. Sixteen normal breast tissues and 119 paraffin-embedded breast cancer tissues were used to analyze YAP1 expression via IHC. We selected 51 samples of postoperative breast cancer tissues resected between 2015 and 2021 from the Harbin Medical University Cancer Hospital to analyze the association of YAP1 and NEDD4L expression. Tissue microarrays (constructed using 362 breast cancer and 28 normal paraffin-embedded breast tissues) were used for IHC staining; survival data were obtained from the corresponding patients. Two clinical survival outcome endpoints were chosen for the endpoint analysis: overall survival (OS, from the date of surgery to the date of death or last follow-up) and disease-free survival (DFS, from the date of surgery to the date of recurrence or metastasis). The relationships of total YAP1, cytoplasmic YAP1, and nuclear YAP1 expression with breast cancer survival were analyzed. The use of human tissues was approved by the Ethics Committee of Harbin Medical University Cancer Hospital (KY2016-13, KY2018-09, KY2022-27), and written informed consent was obtained from all patients.
2.5 Detection of autophagy
When detecting the expression levels of autophagy markers (LC3 II and p62 proteins), EBSS was used to starve cells to induce autophagy, and the late autophagy inhibitor CQ was used to inhibit the fusion of autophagosomes and lysosomes, both of which were added 6 h before collecting cells. Adenovirus of mRFP-GFP-LC3 was used to detect autophagic flux. IF was used to detect punctate aggregation of GFP-LC3. Transmission electron microscopy (TEM) was used to image autophagic structures in breast cancer cells and tissues.
2.6 WB analysis
WB analyses were performed as previously described [25]. Cells or tissue samples were lysed using radio-immunoprecipitation assay (RIPA) lysis buffer (AR0102, Boster) and the protease inhibitor phenylmethylsulfonyl fluoride (PMSF, AR1178, Boster). After sonication, samples were lysed on ice for 30 min, then centrifuged at 13,500 × g for 15 min at 4°C. The protein concentration in the supernatant was determined using a Pierce™ BCA Protein Assay Kit (23225, Thermo Fisher Scientific, Waltham, MA, USA) and denatured using sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) protein loading buffer (AR1112, Boster). Gel electrophoresis was performed using a gel kit (PG112, PG113, EpiZyme, Shanghai, China), and the protein was transferred to a polyvinylidene fluoride (PVDF) membrane (3010040001, Roche, Shanghai, China). Enhanced chemiluminescence (ECL) buffer (P2300, New Cell & Molecular Biotech, Suzhou, Jiangsu, China) was used for color development.
2.7 IHC staining
After deparaffinization, antigen retrieval, and the addition of primary antibody overnight at 4°C, pathological tissue sections were treated with the appropriate secondary antibody and stained with 3,3′-diaminobenzidine (DAB) the following day. After counterstaining with hematoxylin, the sections were mounted. Three fields were randomly selected, and samples were scored based on the percentage of positive cells and the staining intensity as follows: 0 (no staining), 1 (light yellow), 2 (yellow-brown), or 3 (brown). The proportion of positively stained tumor cells in a given region was scored as 0 (0-25% positive tumor cells), 1 (25%–50% positive tumor cells), 2 (50%–75% positive tumor cells), or 3 (75%–100% positive tumor cells). The final staining score for each sample was equal to the staining intensity plus the proportion of positive staining. The median was selected as the cutoff value. A staining score greater than or equal to the median was considered as high expression, and lower than the median was considered as low expression.
2.8 CCK-8 assay
Cells were cultured as described above, trypsinized, and passaged in 96-well plates with 3,000-5,000 cells/well. At 0, 24, 48, and 96 h after the cells adhered, the culture medium was discarded. A mixture of culture medium and CCK-8 solution (GK10001, GlpBio, Montclair, CA, USA) (9:1) was then added (100 µL/well). After incubation at 37°C for 2 h, a microplate reader was used to measure the optical density at 450 nm (OD450).
2.9 EdU incorporation assay
Cells were cultured as described above, trypsinized, and passaged in a 96-well plate with 5,000 cells/well. At 48 h after the cells adhered, the EdU incorporation assay was performed as instructed by the manufacturer (YF®594 Click-iT EdU Imaging Kits, C6015, US Everbright, Suzhou, Jiangsu, China). In brief, culture medium (100 µL/well) containing 50 µmol/L EdU was added to the plate and incubated at 37°C for 3 h to label the DNA. After incubation, cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% phosphate-buffered saline (PBS)-Triton X-100 (1139ML100, Biofroxx, Shanghai, China), then the prepared reaction mixture containing fluorescent dye was added, and plates were incubated at 25°C in the dark. The nuclei were then stained with Hoechst 33342 for 30 min.
2.10 Quantitative real-time PCR (qRT-PCR)
RNA was extracted from cultured cells with TRNzol (DP424, TIANGEN Biotech, Beijing, China) following the manufacturer's protocol. After the concentration of RNA was determined, cDNA was synthesized from 1 µg of RNA per sample using the FastKing gDNA Dispelling RT SuperMix (KR118, TIANGEN Biotech). Amplification was performed using Talent qPCR PreMix containing SYBR Green (FP209, TIANGEN Biotech). PCR was performed using the following conditions: an initial denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 60°C for 60 s. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the internal reference gene for normalization. Relative expression was calculated using the 2−𝛥𝛥CT method [26]. The primer sequences are shown in Supplementary Table S1.
2.11 Cell apoptosis and necrosis staining
Cells were cultured as described above, trypsinized, and passaged in a 96-well plate with 5,000 cells/well. After the cells adhered, the cell apoptosis and necrosis staining was performed using the Apoptosis and Necrosis Assay Kit (C1056, Beyotime, Shanghai, China) as instructed by the manufacturer. The mixture was prepared by the addition of 5 µL Hoechst33342 and 5 µL propidium iodide (PI) to 800 µL cell staining buffer and mixed well. The mixture was then added to a 96-well plate (100 µL/well), incubated at 4°C for 30 min, and washed once with PBS before acquiring photographs.
2.12 Protein-protein interaction (PPI) network construction
To explore the mechanism by which YAP1 promoted AP formation, we generated a PPI network using the autophagy gene set published by Wang et al. [27]. The STRING database (https://cn.string-db.org) was used to identify interactions between YAP1 and the autophagy gene set. After downloading the interaction results (Supplementary Table S2), Cytoscape software v3.6.0 (Cytoscape Consortium, New York, NY, USA) was used to visualize the network.
2.13 Molecular docking
The crystal structures of YAP1 and the carboxy terminus of CHMP2B were downloaded from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (https://www.rcsb.org/). The crystal structure of YAP1 was isolated from the 5OAQ crystal structure, and the carboxy-terminal crystal structure of CHMP2B was isolated from the 2JQK crystal structure. The structure of VPS4B (AF-O75351-F1) was downloaded from the AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk/). Molecular docking was performed with the ZDOCK SERVER (https://zdock.umassmed.edu/) and visualized with PyMol v2.5.0 (Schrodinge LLC, New York, NY, USA).
2.14 IF staining
IF staining was performed as previously described [28]. In brief, cells were cultured and seeded in small confocal dishes with 5 × 104 cells/dish. After transfection with plasmids for 48 h or EGCG (an agonist of cytoplasmic YAP1 localization) treatment for 6 h, cells were fixed with 4% paraformaldehyde (BL539A, Biosharp Life Sciences, Beijing, China), permeabilized with 0.5% PBS-Triton X-100, and blocked with 10% goat serum (AR0009, Boster). Cells were incubated with primary antibody overnight at 4°C, then incubated with the corresponding secondary antibody (Dylight 488/594 Goat Anti-Mouse/Rabbit IgG, A23210/A23320, Abbkine, Wuhan, Hubei, China) for 1 h at 25°C in the dark the following day. Nuclei were stained with 4',6-diamidino-2-phenylindole (DAPI, AR1176, Boster), after which anti-fluorescence quenching mounting medium (AR0036, Boster) was added. Cells were then imaged with Zeiss LSM 800 Confocal Laser Scanning Microcopy (Carl Zeiss AG, Oberkochen, Germany).
2.15 TEM
Cells were cultured as described above, digested with ethylene diamine tetraacetic acid (EDTA)-free trypsin (PYG0065, Boster), and centrifuged at 25°C and 3,000 × g for 5 min. Pelleted cells or fresh tissue sections were rinsed with PBS and fixed overnight with 2.5% glutaraldehyde. After ethyl alcohol dehydration, resin infiltration and embedding, dicing, and ethyl alcohol lead citrate staining [29], photos were obtained using a TEM (Carl Zeiss AG).
2.16 Co-IP
Protein A/G Magnetic Beads (HY-K0202, MedChemExpress) were washed four times with 0.5% PBS-Triton X-100. The antibody solution was then added, and the magnetic beads and antibodies were incubated for 1 h at 25°C on a flip mixer. The antibody-conjugated magnetic beads were washed four times with 0.5% PBS-Triton X-100. Prepared protein supernatant was then added, and the antibodies and proteins conjugated to the magnetic beads were incubated for 2 h at 4°C on a flip mixer. After washing with 0.5% PBS-Triton X-100, 1 × SDS-PAGE protein loading buffer was added; samples were denatured by heating, and proteins were measured by WB.
2.17 Silver staining
Silver staining was performed as previously described [30] using the Fast Silver Stain Kit (P0017S, Beyotime). Briefly, after electrophoresis, the protein gel was fixed overnight with a solution containing ethanol and acetic acid following the manufacturer's instructions. After washing with 30% ethanol followed by Milli-Q grade pure water, the silver dye sensitization solution was added, then the silver solution was used. Samples were washed with Milli-Q grade pure water, then the silver dye color developing solution was used to develop the color. Images were taken with a SONY DSC-HX99 photo camera (Tokyo, Japan) after the color development was halted.
2.18 Protein stability assay and ubiquitin assay
The cells were cultured as described above, and when the cell density was 60%-70%, CHX (100 µg/mL) was added for 8, 10, and 12 h, then the cells were collected, and the protein stability of YAP1 was detected by WB.
The Flag-ub plasmid was used to overexpress ubiquitin, then cells were treated with the proteasome inhibitor MG132 (20 µmol/L) for 6 h before harvesting. Anti-Flag-ub antibody was used for co-IP, and ubiquitin was detected by WB.
2.19 Nuclear and cytoplasmic protein extraction
The Subcellular Structure Nuclear and Cytoplasmic Protein Extraction Kit (AR0106, Boster) was used as instructed by the manufacturer. Cells were spheroidized under hypotonic conditions (lower than intracellular osmotic pressure), then the cytoplasmic proteins in the supernatant were isolated by centrifugation (4°C, 16,000 × g for 5 min). Nuclear proteins were extracted with a high-salt nuclear protein extraction reagent. Specifically, cytoplasmic protein extraction reagent A was added to the cell pellet, and samples were shaken to mix, then incubated on ice for 10-15 min. Cytoplasmic protein extraction reagent B was then added, and samples were shaken to mix, incubated on ice for 1 min, and centrifuged at 4°C and 16,000 × g for 5 min. Cytoplasmic proteins were present in the supernatant, which was aspirated. Nucleoprotein extraction reagent was added, samples were shaken to mix, and then incubated on ice for 40 min with shaking for 15 s every 10 min. Finally, samples were centrifuged at 4°C and 16,000 × g for 5 min and aspirated to obtain the nucleoproteins. Cytoplasmic proteins and nucleoproteins were detected by WB.
2.20 Xenograft model
Female BALB/c nude mice (4-5 weeks old, 18-20 g) were purchased from Vital River (Shanghai, China). All animal experiments were performed according to the guidelines approved by the Institutional Animal Care and Use Committee (IACUC) of Harbin Medical University (GB/T 35892-2018) and the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals. MDA-MB-231 cells (5 × 106 in 0.2 mL PBS:Matrigel [3:1] mix solution) were injected subcutaneously into the backs of 24 nude mice. After palpable tumor formation, mice were divided into 4 subgroups of 6 mice each. EGCG (100 mg/kg, an agonist of cytoplasmic YAP1 localization) was administered by gavage, and XMU-MP-1 (1 mg/kg, an inhibitor of cytoplasmic YAP1 localization) was administered by intraperitoneal injection every day for 30 days. The long (L) and short (W) diameters of the tumors were measured with vernier calipers every 2 days, and the tumor volume was estimated using the formula (L × W2) / 2. After 30 days of treatment, all mice were euthanized by cervical dislocation, the tumors were removed, and the mass of each tumor was then weighed and recorded.
2.21 Bioinformatics analysis
Breast cancer YAP1 mRNA expression datasets were downloaded from the Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/) and cBioPortal databases (https://www.cbioportal.org/). Differential expression of YAP1 in normal breast and breast cancer tissues was analyzed using the ‘beeswarm’ package, and survival maps were generated using the ‘survival’ and ‘survminer’ packages by RStudio v1.4.1717 (RStudio Inc., Boston, MA, USA). Two clinical survival outcome endpoints were chosen for the endpoint analysis: OS (from the date of diagnosis to the date of death or last follow-up) and relapse-free survival (RFS, from the date of diagnosis to the date of recurrence).The survival maps of P46937 and 224894_at datasets were downloaded from the Kaplan-Meier online database (https://kmplot.com/analysis/). OS and RFS were chosen for the endpoint analysis. The representative IHC staining images of normal and breast cancer tissues in Supplementary Figure S1A were downloaded from the Human Protein Atlas (HPA) (https://www.proteinatlas.org/). HPA is a freely available interactive resource portal, offering the possibility to explore the tissue-elevated proteomes in tissues and organs and to obtain information on the subcellular localization of proteins [31, 32]. The pictures used were obtained from https://www.proteinatlas.org/ENSG00000137693-YAP1/pathology/breast+cancer#imid_19107801, and are available from v21.0.proteinatlas.org. To identify the small-molecule compounds related to the Hippo pathway, we used the online database Natural Product Activity & Species Source Database (NPASS, http://bidd.group/NPASS/) to download the related compounds with breast cancer cell lines MDA-MB-231, MDA-MB-468, and MCF7. After taking the intersection, the STITCH database (http://stitch.embl.de/) was used to identify interactions between these compounds and the Hippo gene set. After downloading the interaction results, Cytoscape software v3.6.0 (Cytoscape Consortium) was used to visualize the network.
To identify EGCG related to the Hippo pathway, the expression profile GSE56245 was downloaded from the Gene Ommnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/), and the ‘limma’ package in R was used to screen differentially expressed genes (DEGs). The screening threshold was P < 0.01. Weighted gene co-expression analysis (WGCNA) was used to construct a co-expression network. Then, six gene modules were constructed according to the degree of co-expression, namely MEblue, MEyellow, MEgreen, MEbrown, MEturquoise and MEred, and the genes in these modules were analyzed by correlation clustering to produce a heat map of the correlation. We then performed a Kyoto Encyclopedia of Genes and Genomes (KEGG) biochemical pathway enrichment analysis using the DAVID online database (https://david.ncifcrf.gov/tools.jsp). Finally, the downloaded data from DAVID was visualized using the ‘ggplot2’ package of RStudio v1.4.1717.
To obtain YAP1-related ubiquitin ligases, we used the online database UbiBrowser (http://ubibrowser.bio-it.cn/ubibrowser_v2/) to predict and downloaded the results. UbiBrowser 2.0 is an integrated database for predicted human proteome-wide E3-substrate interactions [33, 34]. As a discovery tool, UbiBrowser 2.0 contributes to the study of protein ubiquitination and the development of drug targets for complex diseases.
2.22 Cox proportional hazards analyses and nomogram construction
Univariate and multivariate COX survival analyses were performed using SPSS v23.0 (SPSS Inc., Chicago, IL, USA). With the support of RStudio v1.4.1717, we used the ‘rms’ and ‘survival’ packages to build the nomogram model, and the ‘timeROC’ package to produce the receiver operating characteristic (ROC) curve and calculate the area under the curve (AUC).
2.23 Statistical analysis
The data from in vitro experiments and from nude mouse xenograft experiments were analyzed as the mean ± standard deviation of at least three and six independent experiments, respectively. Student's t-test and one-way analysis of variance (ANOVA) were used to determine statistically significant differences between groups at each time point. Survival curves were drawn using the Kaplan-Meier method and compared with a log-rank test. P < 0.05 was considered statistically significant. Statistical analysis was performed using the SPSS v23.0 and GraphPad Prism v8.0 (GraphPad Software Inc., San Diego, CA, USA).
3 RESULTS
3.1 Cytoplasmic YAP1 inhibited proliferation of breast cancer cells
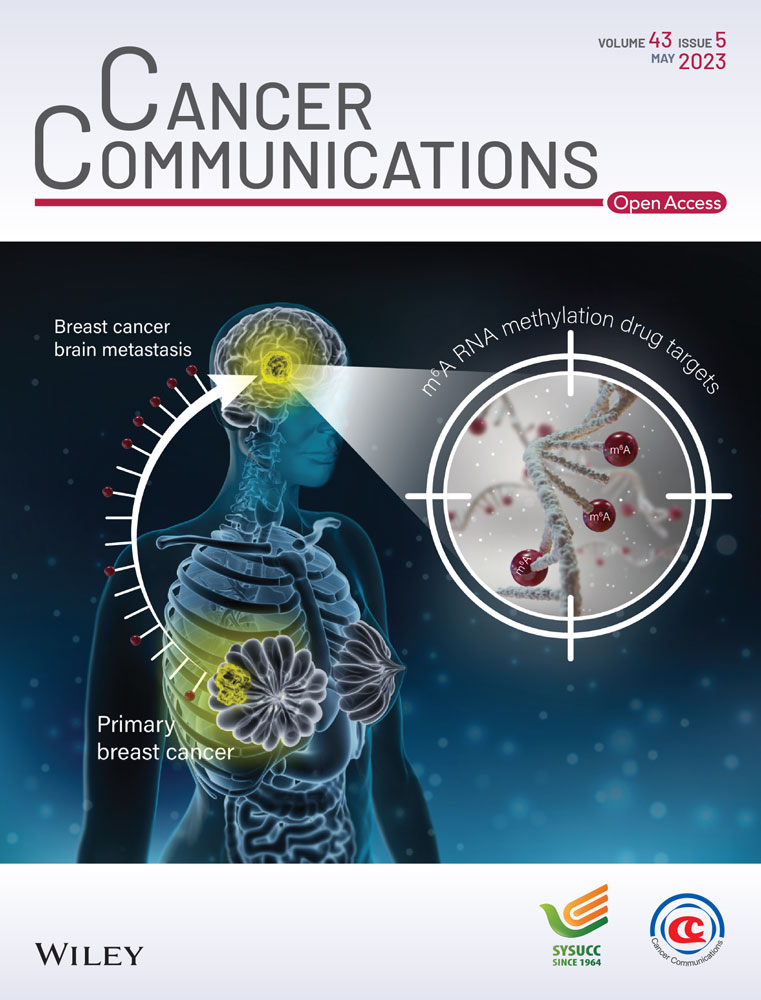
Using IHC staining, we identified 16 normal breast tissues and 119 breast cancer tissues from two patient cohorts from the Harbin Medical University Cancer Hospital. YAP1 was expressed at significantly lower levels in breast cancer tissues than in normal breast tissues, and was mainly expressed in the cytoplasm of breast cancer tissues (Figure 1A-B). In the breast cancer mRNA expression datasets from TCGA, we also found that YAP1 expression was significantly lower in breast cancer tissues than in normal breast tissues (Figure 1C). Six pairs of breast cancer tissues and adjacent normal tissues were selected for WB analysis, which showed that YAP1 expression was lower in breast cancer tissues than in adjacent tissues (Figure 1D). We obtained representative IHC staining images of normal and breast cancer tissues from three patients using four YAP1 antibodies from HPA. All four YAP1 antibodies stained lighter in breast cancer tissues than in normal breast tissues (Supplementary Figure S1A-B). We also used data from three databases to analyze the relationship between YAP1 expression and breast cancer survival. The results of this analysis were inconsistent. High YAP1 protein expression was associated with shorter RFS at the P46937 dataset and high YAP1 mRNA expression was associated with longer overall survival (OS) at the 224894_at dataset from the Kaplan-Meier online database (Supplementary Figure S1C-D). The breast cancer mRNA dataset from TCGA showed that high YAP1 expression was associated with shorter OS (Supplementary Figure S1E), whereas the breast cancer mRNA datasets from cBioPortal showed that high YAP1 expression was associated with longer OS (Supplementary Figure S1F). Using data from TCGA, we analyzed associations between the expression of YAP1 targets (CTGF and CYR61), instead of YAP1 itself, and survival as a measure of actual YAP1 activity in these samples, and found that simultaneous high expression of CTGF and CYR61 was not associated with the survival of breast cancer patients (Supplementary Figure S1G). A recent study showed that YAP1 suppressed estrogen receptor-positive (ER+) breast cancer by inhibiting the expression of estrogen receptor 1 (ESR1) via vestigial like family member 3 (VGLL3) [35]. We therefore explored the association between YAP1 mRNA expression and survival in different molecular subtypes of breast cancer (ER+, human epidermal growth factor receptor 2-positive [HER2+], ER−HER2−) from cBioPortal. We found that YAP1 mRNA expression did not differ between ER+ and HER2+ breast cancers, but was lower in ER+ and HER2+ breast cancers than in ER−HER2− (Supplementary Figure S1H). High YAP1 expression was associated with longer OS in ER+ breast cancer, whereas no association was found in other subtypes (Supplementary Figure S1I-J). Based on these results, the role of YAP1 in breast cancer survival remains unclear and requires further study.
YAP1 mRNA and protein expression levels were measured by qRT-PCR and WB analysis, respectively, in the normal human mammary epithelial cell line MCF-10A and in eight breast cancer cell lines (Figure 1E-F). The gene and protein expression patterns of these cell lines were inconsistent. Because proteins are the main drivers of biological activities, protein levels may provide a more direct and accurate understanding of biological processes than gene expression [36, 37]. The breast cancer cell lines MDA-MB-231 (with high YAP1 protein expression), MCF7 and SKBR3 (both with low YAP1 protein expression) were selected for further study. First, MCF7 and SKBR3 cells were transfected with a YAP1-overexpression plasmid, and MDA-MB-231 cells were transfected with YAP1-siRNAs (siYAP1#1, siYAP1#2, or siYAP1#3), siYAP1#2 had the highest knockdown efficiency (Supplementary Figure S1K). IF showed that YAP1 was more abundant in the cytoplasm after transient overexpression of YAP1 (Supplementary Figure S1L). At the same time, we used a lentiviral system to stably express YAP1 in MCF7 and SKBR3 cells. Based on the results from a CCK-8 assay, MCF7 and SKBR3 cells showed significantly reduced cell proliferation ability after transiently and stably overexpressing YAP1, and MDA-MB-231 cells showed significantly increased cell proliferation ability after YAP1 knockdown (Figure 1G, Supplementary Figure S1M). An EdU incorporation assay showed that the DNA synthesis capacities of MCF7 and SKBR3 cells were significantly weakened after transiently and stably overexpressing YAP1, whereas the DNA synthesis capacity of MDA-MB-231 cells was significantly enhanced after knocking down YAP1 (Figure 1H, Supplementary Figure S1N). We used WB to detect the expression of apoptosis markers (Caspase-3 and PARP) in MCF7 and SKBR3 cells after transient and stable overexpression of YAP1, and the results showed that YAP1 promoted apoptosis (Figure 1I, Supplementary Figure S1O). Cell apoptosis and necrosis was detected by Hoechst 33342 and PI staining after stable YAP1 overexpression in MCF7 and SKBR3 cells, and the results indicated that YAP1 promoted apoptosis and necrosis of breast cancer cells (Supplementary Figure S1P). The results above showed that YAP1 was mainly expressed in the cytoplasm of breast cancer tissues and YAP1-overexpressing breast cancer cells, and that it inhibited proliferation, in contrast to the classical model in which nuclear YAP1 promotes cell proliferation [3, 4].
To better distinguish the subcellular localization of YAP1, we constructed different mutants, including NLS-YAP15SA (promotes the nuclear localization of YAP1 and increases YAP1 transcriptional activity), YAP1S94A (cannot bind to TEADs), and YAP1S127D (cytoplasmic localization of YAP1 phospho-mimic) [4, 5, 38, 39]. CCK-8 assays demonstrated the viability of MCF7 and SKBR3 cells transiently transfected with different mutants of YAP1. YAP1S127D significantly inhibited the proliferation of MCF7 and SKBR3 cells, NLS-YAP15SA promoted SKBR3 cell proliferation, and YAP1S94A inhibited MCF7 cell proliferation (Figure 1J). In the next experiment, we used YAP1S127D to detect DNA synthesis and apoptosis, and also found that YAP1S127D inhibited DNA synthesis and promoted apoptosis in MCF7 and SKBR3 cells (Figure 1K-L). Taken together, the above experimental results indicated that cytoplasmic YAP1 inhibited breast cancer cell proliferation.
3.2 YAP1 promoted AP formation in breast cancer cells
Autophagic cell death is a novel type of programmed cell death. The dynamic process of autophagy includes the activation of autophagy, nucleation of a phagophore assembly site (PAS), elongation of the phagophore (PG), maturation of the AP, and the fusion of the AP and lysosome to form the AL. The entirety of this process is called autophagic flow. The unc-51 like autophagy-activating kinase 1 (ULK1) protein, PI3K kinase, and autophagy-related proteins (ATGs) were activated and recruited to the PAS and PG. LC3, SQSTM1/p62, and substances to be degraded were encapsulated into the PG and closed to form a complete AP; an AL was then formed by fusion of an AP and a lysosome, and encapsulated substances (including LC3 and p62) were degraded. After the formation of the PG, cytoplasmic LC3 was combined with phosphatidylethanolamine (PE) and lipidated to form LC3 II, which was implanted in the inner membrane of the PG; p62, a small vehicle, bound substances to be degraded, then bound to LC3 II in the PG to be degraded. LC3 and p62 were therefore used as markers to detect the occurrence of autophagy and autophagy flow [40-42].
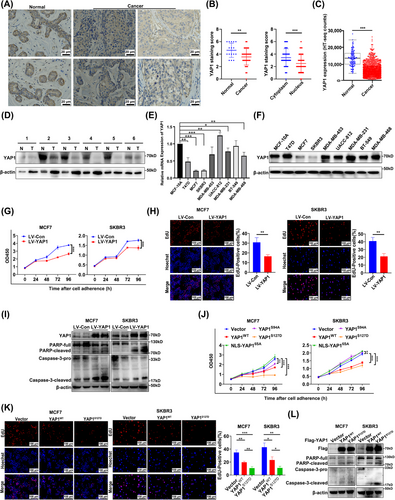
After YAP1 was overexpressed for 48 h in MCF7 and SKBR3 cells, WB analysis showed that levels of the autophagy marker LC3 II were increased and the substrate p62 were decreased. YAP1 expression was confirmed with monoclonal antibodies (normalized to β-actin), indicating that YAP1 increased the basal level of autophagy (Figure 2A). After YAP1 was overexpressed for 24 h, punctate aggregation of GFP-labeled LC3 was increased in MCF7 and SKBR3 cells (Figure 2B).
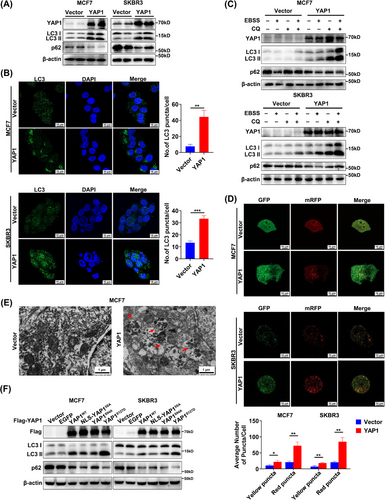
Importantly, the increased levels of LC3 II may represent enhanced AP formation or inhibition of autophagy flow (AL formation) [43]. WB analysis was used to clarify whether the YAP1 overexpression-induced increase in LC3 II activated AP formation or inhibited AP-lysosome fusion (Figure 2C). YAP1 overexpression increased the expression of LC3 II and decreased the expression of p62 in growth media with adequate nutrition. YAP1 overexpression further increased LC3 II expression and decreased p62 expression after starvation with EBSS for 6 h. When AP-lysosome fusion was inhibited by the autophagy inhibitor CQ (20 µmol/L, 6 h) and EBSS was used to induce autophagy, LC3 II and p62 could not be degraded and instead accumulated at very high levels, suggesting that YAP1 promoted AP formation (Figure 2C). We then used an autophagy double standard adenovirus (mRFP-GFP-LC3), which contains an acid-sensitive GFP and an acid-insensitive mRFP, to measure the autophagy flow in response to YAP1 overexpression (Figure 2D). LC3 was labeled with GFP and mRFP, meaning that the AP exhibited yellow fluorescence (GFP+mRFP+). However, because GFP was easily degraded in the acidic lysosomal environment, GFP was degraded when the AP and lysosome fused; red fluorescence (GFP−mRFP+) therefore indicated that an AL had formed and autophagy had flowed smoothly. After 24 h of YAP1 overexpression, both yellow and red fluorescence puncta had significantly increased in number, indicating that YAP1 induced AP formation and that autophagy flow was smooth (Figure 2D). TEM was used to verify that YAP1 overexpression markedly increased the number of autophagic structures in MCF7 cells (Figure 2E). These data thus indicate that YAP1 induced AP formation.
To further clarify the effect of cytoplasmic YAP1 on autophagy, we used different mutants of YAP1 and an EGFP control (excluding the effect of general cell stress) to detect the expression of autophagy markers LC3 and p62; the results suggest that YAP1WT, NLS-YAP15SA, YAP1S94A and YAP1S127D all induced autophagy in MCF7 and SKBR3 cells, and YAP1S127D had the most significant effect (Figure 2F). The experiments strongly demonstrated that cytoplasmic YAP1 promoted autophagy in breast cancer cells.
3.3 YAP1 promoted autophagic death of breast cancer cells
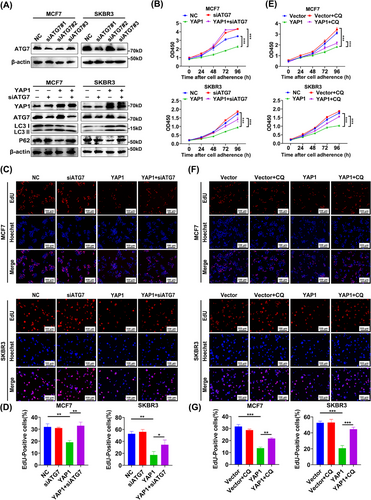
We next examined the effect of YAP1 on breast cancer cell proliferation after inhibition of autophagy to determine whether YAP1-induced proliferation inhibition was dependent upon autophagy. The lipidation process of LC3 occurred through a ubiquitin-like coupling cascade. A ubiquitin E1-like activating enzyme (ATG7) and a ubiquitin E2-like binding enzyme (ATG3) modify LC3 in the cytoplasm, and the ATG12-ATG5-ATG16L1 composite scaffold acted as the ubiquitin ligase E3 to form LC3 II by transferring LC3 from ATG3 to PE [44, 45]. WB analysis of LC3 and p62 showed that, following YAP1 overexpression, knockdown of ATG7 (a key gene for autophagy initiation) by siRNA transfection attenuated the YAP1-induced increase in autophagy marker LC3 II and the decrease in p62 (Figure 3A), indicating suppressed autophagy. CCK-8 and EdU assays showed that ATG7 knockdown partially reversed the anti-proliferation effect of YAP1 (Figure 3B-D). CCK-8 and EdU assays also revealed that the anti-proliferation effect of YAP1 was partially reversed by addition of the autophagy inhibitor CQ (20 µmol/L), which inhibited AP-lysosome fusion (Figure 3E-G). Cell apoptosis and necrosis staining showed that ATG7 knockdown or CQ (20 µmol/L) treatment partially reversed the pro-apoptotic effect of YAP1 in MCF7 cells (Supplementary Figure S2). Together, these data demonstrated that activation of autophagy was a key mechanism by which YAP1 inhibited breast cancer cell proliferation.
3.4 YAP1 promoted assembly of the ESCRT-III complex subunits CHMP2B and VPS4B in the cytoplasm
To explore the specific mechanism by which YAP1 promoted AP formation, we generated a PPI network (Figure 4A) detailing the interactions between YAP1 and autophagy-related genes. We speculated that activation of autophagy by YAP1 may be related to the ESCRT-III complex, namely the subunits VPS25, VPS4B, and CHMP3 [46-48]. To test this hypothesis, qRT-PCR was used to measure the transcription levels of ESCRT-III subunits in YAP1-overexpressing SKBR3 cells and in YAP1-knockdown MDA-MB-231 cells. In contrast to our expectations, YAP1 did not affect expression of genes encoding ESCRT-III subunits (Figure 4B). Whether YAP1 affects assembly of the ESCRT-III complex required future study.
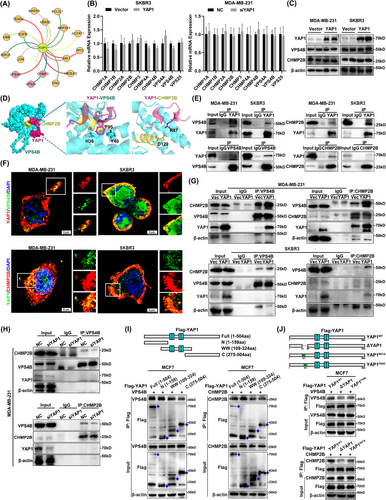
The microtubule interaction and transport (MIT) domain of human VPS4B bound to a conserved sequence motif located at the carboxyl terminus of CHMP1-3 proteins, to which VPS4B and CHMP2B bound weakly [49]. We therefore sought to investigate whether YAP1 could enhance the binding capacity of VPS4B and CHMP2B. WB analysis showed that YAP1 overexpression did not affect protein levels of VPS4B and CHMP2B in MDA-MB-231 and SKBR3 cells (Figure 4C). Molecular docking simulations were then used to study the relevant PPIs (Figure 4D). We first docked YAP1 with VPS4B and the carboxy terminus of CHMP2B, and found that the R87–P99 region of YAP1 was perfectly embedded in the binding regions of VPS4B and CHMP2B. The side chain amino group of YAP1 (R87) formed a hydrogen bond with the hydroxyl group of CHMP2B (D129), and the side chain phenyl ring of YAP1 (F95) formed a π stack with the imidazole of VPS4B (H36) and the phenyl ring of VPS4B (Y40). Therefore, YAP1 acted as an anchor protein to strengthen the binding of VPS4B and CHMP2B (Figure 4D). Co-IP results showed that YAP1 directly interacted with VPS4B and CHMP2B (Figure 4E). Furthermore, co-localization of YAP1-VPS4B and YAP1-CHMP2B was observed in the cytoplasm of MDA-MB-231 and SKBR3 cell lines via IF staining (Figure 4F). To further explore the effect of YAP1 on CHMP2B-VPS4B binding, we examined the interaction between VPS4B and CHMP2B in cell lines with YAP1 overexpressed or knocked down. Co-IP results showed that overexpression of YAP1 enhanced the interaction between VPS4B and CHMP2B in MDA-MB-231 and SKBR3 cells (Figure 4G), whereas YAP1 knockdown suppressed the interaction between VPS4B and CHMP2B in MDA-MB-231 cells (Figure 4H). To determine the specific sequence of YAP1 binding with VPS4B and CHMP2B, we first constructed three truncation mutants of YAP1, including N-terminal (1-159aa), WW domain (109-324aa), and C-terminal (275-504aa), and found that the N-terminal (1-159aa) of YAP1 bound with VPS4B and CHMP2B by co-IP (Figure 4I). Molecular docking predicted that YAP1 (R87-P99) bound with VPS4B-CHMP2B, and the R87-P99 sequence is included in the N-terminus (1-159aa). Therefore, we further deleted the R87-P99 sequence of YAP1 (ΔYAP1) and mutated YAP1 (R87) and YAP1 (F95) to alanine (YAP1R87A, YAP1F95A). Co-IP results showed that ΔYAP1 significantly weakened the binding of YAP1 with VPS4B and CHMP2B, YAP1F95A weakened the binding of YAP1 with VPS4B, while YAP1R87A weakened the binding of YAP1 with CHMP2B (Figure 4J). The R87-P99 sequence of YAP1 could act as an anchor sequence to bind with VPS4B and CHMP2B at the same time, making the binding of two more tight. In summary, cytoplasmic YAP1 promoted AP formation by binding with VPS4B and CHMP2B to promote ESCRT-III assembly.
3.5 EGCG enabled retention of YAP1 in the cytoplasm by activating the Hippo pathway and promoted autophagy of breast cancer cells
The experiments described above preliminarily demonstrated that cytoplasmic YAP1 promoted autophagic death of breast cancer cells by promoting assembly of the ESCRT-III complex subunits CHMP2B-VPS4B. Researchers have previously developed inhibitors to prevent translocation of cytoplasmic YAP1 into the nucleus as an anti-tumor treatment [50, 51]. Natural drugs have attracted a great deal of attention due to the advantages of easy access and low toxicity. We therefore considered using natural small-molecule compounds to increase YAP1 localization in the cytoplasm (rather than to alter total YAP1 levels). This would clearly demonstrate that cytoplasmic YAP1 promoted autophagic death of breast cancer cells. Activation of the Hippo pathway was crucial to regulating the subcellular localization of YAP1; when the Hippo pathway was activated, the downstream classic molecule YAP1 was phosphorylated and could not enter the nucleus, and thus remained in the cytoplasm [3, 10, 52, 53]. We used NPASS to screen 48 natural small-molecule compounds related to the breast cancer cell lines MDA-MB-231, MDA-MB-468, and MCF7 (Supplementary Figure S3A). The interactions of these molecules with genes in the Hippo pathway were studied using the STITCH database; 26 small-molecule compounds related to the Hippo pathway were identified (Supplementary Table S3). The 15 small-molecule compounds that interacted with the most genes in the Hippo pathway were shown in Figure 5A. EGCG, the compound with the sixth highest number of nodes, had been previously confirmed to activate autophagy in tumor cells [54, 55], but the mechanism of this action remained unclear. Expression profiles of breast cancer MCF7 cells treated with ethanol (EtOH), EGCG, estrogen (E2), and EGCG + E2 were downloaded from the GEO database (GSE56245), and we screened 4342 DEGs between EGCG, EGCG + E2 groups and the EtOH, E2 groups. WGCNA was used to construct a co-expression network. We clustered DEGs into six modules based on the degree of co-expression: blue (MEblue), green (MEgreen), yellow (MEyellow), brown (MEbrown), sky blue (MEturquoise), and red (MEred) (Supplementary Figure S3B). After clustering the DEGs into those six modules, we re-clustered the genes within each module to more accurately model the degree of co-expression. We found that the co-expression correlation of genes was highest in the MEturquoise module (Supplementary Figure S3C). We then associated the sample treatment groupings (EtOH, E2, EGCG, EGCG + E2) with the six co-expression modules; consistent with the results described above, the genes in the MEturquoise module had the highest correlation (Supplementary Figure S3D). We therefore selected the MEturquoise module for further analysis. We performed a KEGG biochemical pathway enrichment analysis using the DAVID database, and selected the 10 most significantly enriched pathways (including Hippo) (Figure 5B, Supplementary Table S4). We examined the effect of EGCG on components of the Hippo pathway and the expression of YAP1 target genes (CTGF and CYR61) using WB analysis. EGCG treatment resulted in an increase in p-MST1 (Thr183), p-MOB1A (Thr35), and p-YAP1 (Ser127) without affecting total YAP1 protein expression. EGCG treatment significantly decreased YAP1 levels in the nucleus and increased p-YAP1 in the cytoplasm, while EGCG treatment reduced the expression of CTGF and CYR61 (Figure 5C-D). These results suggested that EGCG promoted localization of YAP1 to the cytoplasm.

We next examined the effect of EGCG on autophagy in breast cancer cells. CCK-8 assays showed that EGCG inhibited MCF7 and MDA-MB-231 cell viability (Supplementary Figure S3E). EdU assays showed that EGCG (50 µg/mL, 48 h) inhibited DNA synthesis in MCF7 and MDA-MB-231 cells (Supplementary Figure S3F). WB analysis demonstrated that EGCG-treated MCF7 and MDA-MB-231 cells showed increases in the autophagy marker LC3 II and decreases in p62 (Figure 5E-F). After EGCG treatment (50 µg/mL, 48 h), GFP-labeled LC3 showed punctate aggregation in MCF7 and MDA-MB-231 cells (Figure 5G). The autophagic structures of MCF7 and MDA-MB-231 cells treated with EGCG (50 µg/mL, 48 h) were visualized with TEM (Figure 5H). These experimental results proved that EGCG promoted the retention of YAP1 in the cytoplasm and autophagy of breast cancer cells.
3.6 Cytoplasmic retention of YAP1 promoted autophagic death of breast cancer cells after EGCG treatment
The experiments conducted to this point showed that EGCG could promote autophagy in breast cancer cells. We further used CQ, a late autophagy inhibitor, to determine whether EGCG could also promote autophagic death of breast cancer cells. CCK-8 and EdU assays showed that CQ treatment (20 µmol/L) attenuated the inhibitory effect of EGCG on the proliferation of breast cancer cells (Figure 6A-B). As predicted, EGCG promoted autophagic death in breast cancer cells.
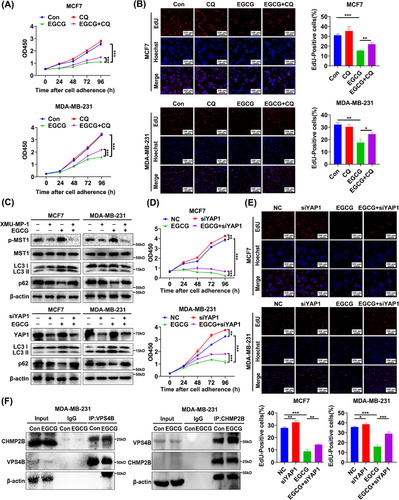
MST1 kinase is a core upstream component of the Hippo pathway. XMU-MP-1 inhibits the activity of MST1 kinase and thus inhibits activation of the Hippo pathway. Under these circumstances, YAP1 could not be phosphorylated, was retained in the cytoplasm, and was therefore activated in the nucleus [56-58]. Inhibiting MST1 activity by treatment with XMU-MP-1 (8 µmol/L, 6 h) weakened the effects of EGCG on the autophagy markers LC3 II and p62, as did YAP1 knockdown (Figure 6C). CCK-8 and EdU assays also indicated that the anti-proliferation effect of EGCG was partially reversed by YAP1 knockdown (Figure 6D-E). This suggest that retention of YAP1 in the cytoplasm was necessary for EGCG to promote autophagic death of breast cancer cells. Surprisingly, EGCG treatment enhanced the interaction between the ESCRT-III complex subunits CHMP2B and VPS4B in MDA-MB-231 cells (Figure 6F). Collectively, these results further clarified that cytoplasmic YAP1 promoted autophagic death of breast cancer cells.
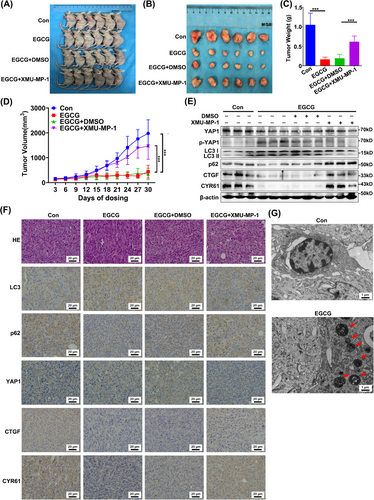
3.7 Cytoplasmic YAP1 promoted autophagic death of breast cancer cells in vivo
We next used EGCG to retain YAP1 in the cytoplasm in vivo to study the effect of cytoplasmic YAP1 on the growth of xenograft tumors in mice. Tumor volume and tumor mass were smaller in mice treated with EGCG (100 mg/kg) than in untreated mice or those treated with the Hippo pathway inhibitor XMU-MP-1 (1 mg/kg) and EGCG (100 mg/kg) (Figure 7A-D). These results indicated that YAP1 retention in the cytoplasm was associated with decreased tumor size, whereas treatment with XMU-MP-1 inhibited retention of YAP1 in the cytoplasm, which was associated with increased tumor size. This demonstrated that cytoplasmic YAP1 inhibited the growth of xenograft tumors. WB analysis showed that p-YAP1 and LC3 II levels were increased, while p62, CTGF and CYR61 levels were decreased in the EGCG-treated group, which were reversed by EGCG + XMU-MP-1 co-treatment (Figure 7E). The IHC staining results confirmed that YAP1 was retained in the cytoplasm in the EGCG-treated group and that the inhibitor XMU-MP-1 caused accumulation of YAP1 in the nucleus; additionally, LC3 levels were increased and p62, CTGF, CYR61 levels were decreased in the EGCG group, and the opposite effect occurred in the EGCG + XMU-MP-1 co-treatment group (Figure 7F). Furthermore, TEM showed that there was a significant increase in autophagic structures in the EGCG-treated group (Figure 7G). Taken together, these in vivo data strongly supported the hypothesis that cytoplasmic YAP1 promoted autophagic death of breast cancer cells.
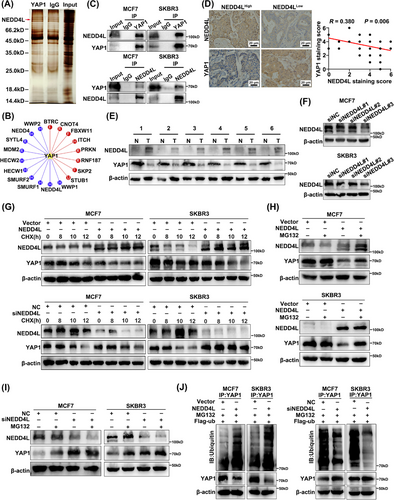
3.8 NEDD4L mediated YAP1 ubiquitination and degradation
The ubiquitin-proteasome pathway was important for maintenance of appropriate cellular YAP1 levels; YAP1 was easily ubiquitinated and degraded by the beta-transducin repeat containing E3 ubiquitin protein ligase (β-TRCP), CCR4-NOT core ubiquitin-protein ligase subunit (Not4), and other ubiquitin ligases in the cytoplasm [59, 60]. The deubiquitination enzymes ubiquitin specific peptidase 10 (USP10), ubiquitin specific peptidase 47 (USP47), OUT deubiquitinase, ubiquitin aldehyde binding 2 (OTUB2), and eukaryotic translation initiation factor 3 subunit H (EIF3H) deubiquitinated YAP1 to maintain protein stability [61-63]. We next identified proteins involved in regulating the stability of YAP1 in the ubiquitin protease system of breast cancer cells. MDA-MB-231 cell lysates were immunoprecipitated with anti-YAP1 antibody, silver-stained with gel electrophoresis, and identified by mass spectrometry. The ubiquitin ligase NEDD4L was identified as a potential interactor with (and degrader of) YAP1 (Figure 8A). UbiBrowser was also used to predict ubiquitin ligases that acted on YAP1. A total of 18 E3 ubiquitin ligases were predicted as candidates for YAP1. Among them, 8 in red had been verified as E3 ubiquitin ligases of YAP1 [59, 64-69], meaning the other 10 in blue were predicted but unconfirmed as E3 ubiquitin ligases of YAP1. NEDD4L had the second highest predicted score among the unconfirmed E3 ubiquitin ligases (Figure 8B). As expected, the interaction between YAP1 and NEDD4L was confirmed in MCF7 and SKBR3 cells via co-IP (Figure 8C). IHC staining showed that YAP1 expression was negatively correlated with NEDD4L expression (Figure 8D). Using 6 matched pairs of cancerous and adjacent normal breast tissues, WB analysis showed that NEDD4L and YAP1 expression levels were negatively associated in breast cancer tissues (Figure 8E). The knockdown efficiency of siNEDD4Ls (siNEDD4L#1, siNEDD4L#2, siNEDD4L#3) were detected, and siNEDD4L#3 was selected for subsequent experiments (Figure 8F). NEDD4L was then overexpressed or knockdown in MCF7 and SKBR3 cells, which were subsequently treated with CHX (100 µg/mL, 6 h), an inhibitor of protein synthesis. NEDD4L overexpression decreased the half-life of YAP1 and NEDD4L knockdown prolonged the half-life of YAP1 (Figure 8G). MCF7 and SKBR3 cells overexpressing NEDD4L were also exposed to the proteasome inhibitor MG132 (20 µmol/L, 6 h), which partially blocked YAP1 degradation (Figure 8H). Knocking down NEDD4L decreased YAP1 degradation (Figure 8I). YAP1 ubiquitination was significantly enhanced by NEDD4L overexpression in MCF7 and SKBR3 cells transfected with Flag-ub; knocking down NEDD4L significantly weakened this effect (Figure 8J). These results suggested that the ubiquitin ligase NEDD4L degraded YAP1, and that YAP1 levels in the cytoplasm could be maintained by interfering with NEDD4L expression. The CCK-8 and EdU assays showed that overexpression of NEDD4L increased the cell viability and DNA replication of MCF7 and SKBR3 cells, promoted the proliferation, and this phenomenon was suppressed by overexpression of YAP1 (Supplementary Figure S4A-B). Similarly, knockdown of NEDD4L inhibiting cell proliferation was restored by knockdown of YAP1 (Supplementary Figure S4C-D). The above results indicated that NEDD4L degraded cytoplasmic YAP1 and inhibited the function of cytoplasmic YAP1.
3.9 High cytoplasmic YAP1 expression was positively correlated with longer survival in breast cancer patients
To analyze the effect of cytoplasmic YAP1 on breast cancer patient prognosis, we performed IHC staining on a tissue microarray containing 362 breast cancer samples and 28 normal breast tissue samples. Total YAP1, cytoplasmic YAP1, and nuclear YAP1 levels were scored for each sample. After survival follow-up for the 362 breast cancer patients, we found that total YAP1 expression did not affect OS or DFS in breast cancer patients. However, high cytoplasmic YAP1 expression and low nuclear YAP1 expression were associated with longer OS and DFS in breast cancer patients (Figure 9A). In addition, we molecularly typed all patients and found that total YAP1 was not differentially expressed in different molecular subtypes (ER+, HER2+, ER−HER2−) (Supplementary Figure S5A), and total YAP1 was positively associated with shorter OS in ER+ breast cancer patients (Supplementary Figure S5B-C).
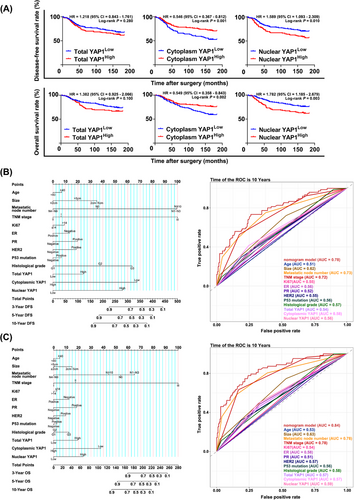
We next performed univariate and multivariate Cox proportional hazards analysis to determine the relationship between 13 clinical features (including total YAP1, cytoplasmic YAP1, and nuclear YAP1 levels) and the survival of breast cancer patients. Cytoplasmic YAP1 expression and nuclear YAP1 expression were independent prognostic factors for breast cancer patient DFS. Total YAP1, cytoplasmic YAP1, and nuclear YAP1 were independent prognostic factors for breast cancer patient OS (Table 1). According to the National Comprehensive Cancer Network guidelines and Chinese Society of Clinical Oncology guidelines, 13 variables were selected to establish the prognostic prediction nomograms for breast cancer [70, 71]. The C-index (0.76 for DFS and 0.80 for OS) and the AUC (0.78 for DFS and 0.84 for OS) indicated satisfactory discriminative ability of the nomograms (Figure 9B-C). In summary, clinical breast cancer data suggested that high cytoplasmic YAP1 levels were beneficial for survival in breast cancer patients; incorporating cytoplasmic YAP1 and nuclear YAP1 expression together in the assessment of survival time in breast cancer patients yielded more accurate results than using total YAP1 expression alone. Finally, the mechanism diagram for the whole article is shown in Figure 10.
| DFS | OS | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate Analysis | Multivariate Analysis | Univariate Analysis | Multivariate Analysis | |||||||||
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Age (≥40 vs. <40) | 1.099 | 0.687-1.758 | 0.694 | 1.103 | 0.603-2.018 | 0.749 | 1.255 | 0.737-2.138 | 0.404 | 1.181 | 0.598-2.333 | 0.631 |
| Size | ||||||||||||
| 2 cm-5 cm vs. ≤ 2cm | 1.823 | 1.221-2.724 | 0.003 | 1.656 | 0.934-2.935 | 0.084 | 2.063 | 1.317-3.231 | 0.002 | 1.671 | 0.878-3.179 | 0.118 |
| >5 cm vs. ≤ 2cm | 5.212 | 2.753-9.869 | <0.001 | 1.331 | 0.464-3.816 | 0.595 | 5.775 | 2.919-11.427 | <0.001 | 1.022 | 0.346-3.013 | 0.969 |
| Metastatic node number | ||||||||||||
| N1-N3 vs. N0 | 2.247 | 1.385-3.646 | 0.001 | 2.470 | 1.236-4.936 | 0.010 | 2.214 | 1.257-3.901 | 0.006 | 1.469 | 0.672-3.213 | 0.335 |
| N4-N9 vs. N0 | 2.963 | 1.750-5.018 | <0.001 | 0.612 | 0.058-6.474 | 0.684 | 4.155 | 2.357-7.324 | <0.001 | 0.162 | 0.015-1.804 | 0.139 |
| N≥10 vs. N0 | 9.457 | 5.861-15.259 | <0.001 | 2.516 | 0.263-24.09 | 0.423 | 12.030 | 7.112-20.349 | <0.001 | 0.732 | 0.074-7.225 | 0.790 |
| TNM stage | ||||||||||||
| II vs. I | 2.838 | 1.442-5.585 | 0.003 | 0.911 | 0.340-2.438 | 0.853 | 4.176 | 1.646-10.596 | 0.003 | 1.834 | 0.535-6.288 | 0.335 |
| III vs. I | 8.321 | 4.257-16.265 | <0.001 | 3.746 | 0.326-42.99 | 0.289 | 16.254 | 6.519-40.528 | <0.001 | 30.481 | 2.289-405 | 0.010 |
| Ki67 (≥14 vs. <14) | 2.122 | 1.030-4.371 | 0.041 | 1.141 | 0.524-2.482 | 0.739 | 1.917 | 0.884-4.159 | 0.099 | 0.916 | 0.391-2.146 | 0.841 |
| ER (+ vs. -) | 0.738 | 0.497-1.094 | 0.131 | 0.824 | 0.483-1.405 | 0.477 | 0.679 | 0.442-1.042 | 0.077 | 0.650 | 0.361-1.169 | 0.150 |
| PR (+ vs. -) | 0.998 | 0.622-1.599 | 0.992 | 1.329 | 0.694-2.544 | 0.391 | 1.084 | 0.637-1.845 | 0.766 | 1.888 | 0.906-3.936 | 0.090 |
| HER2 (+ vs. -) | 1.395 | 0.893-2.180 | 0.144 | 1.301 | 0.751-2.252 | 0.348 | 1.479 | 0.915-2.389 | 0.110 | 1.390 | 0.764-2.530 | 0.281 |
| P53 mutation (+ vs. -) | 1.499 | 0.992-2.263 | 0.054 | 1.099 | 0.676-1.786 | 0.704 | 1.381 | 0.879-2.168 | 0.161 | 1.088 | 0.638-1.855 | 0.757 |
| Histological grade | ||||||||||||
| G2 vs. G1 | 2.405 | 0.885-6.539 | 0.085 | 1.729 | 0.408-7.332 | 0.457 | 2.695 | 0.852-8.529 | 0.092 | 1.036 | 0.239-4.496 | 0.962 |
| G3 vs. G1 | 4.637 | 1.427-15.068 | 0.011 | 2.498 | 0.471-13.23 | 0.282 | 6.166 | 1.668-22.797 | 0.006 | 1.531 | 0.283-8.290 | 0.621 |
| Total YAP1 (high vs. low) | 1.219 | 0.850-1.747 | 0.282 | 1.415 | 0.822-2.436 | 0.210 | 1.383 | 0.938-2.039 | 0.102 | 1.944 | 1.056-3.577 | 0.033 |
| Cytoplasmic YAP1 (high vs. low) | 0.545 | 0.380-0.781 | 0.001 | 0.382 | 0.227-0.643 | <0.001 | 0.548 | 0.371-0.811 | 0.003 | 0.272 | 0.150-0.494 | <0.001 |
| Nuclear YAP1 (high vs. low) | 1.589 | 1.114-2.268 | 0.011 | 2.210 | 1.304-3.745 | 0.003 | 1.783 | 1.213-2.621 | 0.003 | 2.245 | 1.253-4.020 | 0.007 |
- Note: n = 362. TNM stage was determined based on the 8th edition of AJCC TNM Staging system.
- Abbreviations: YAP1, yes1-associated transcriptional regulator; DFS, disease-free survival; OS, overall survival; HR, hazard ratio; CI, confidence interval.
4 DISCUSSION
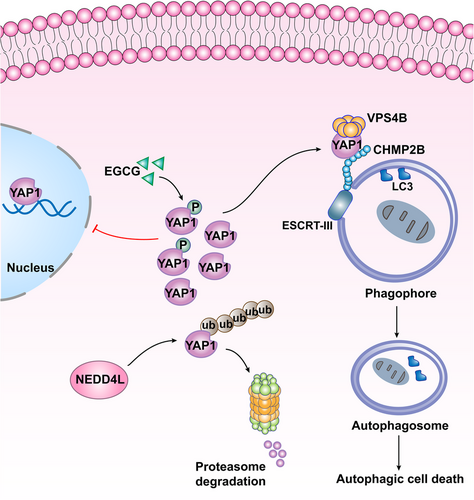
In the present study, we established cytoplasmic mutant YAP1S127D and EGCG-induced YAP1-cytoplasmic model in vitro and in vivo to investigate the role of cytoplasmic YAP1 in ESCRT-III complex and autophagy in breast cancer. We found that cytoplasmic YAP1 led to active autophagy and promoted autophagic death of breast cancer cells through anchoring CHMP2B-VPS4B. Then, we constructed a survival prediction model including cytoplasmic YAP1 using breast cancer tissue chips. Cytoplasmic YAP1 exerted an opposite breast cancer inhibitory effect compared to nuclear YAP1, and was beneficial to the survival of breast cancer patients.
YAP1 was originally found to be involved in regulating organ overgrowth in Drosophila melanogaster, but was later found to be widely activated in human malignancies [3, 72]. It is essential in the formation and growth of many solid tumors and promotes cancer cell proliferation, metastasis, metabolism, stem cell properties, and chemotherapy resistance [73-76]. Research into whether YAP1 can be used as a target for precision tumor therapy has never been completed. We demonstrated that YAP1 expression was significantly lower in breast cancer tissues than in normal breast tissues, consistent with published literature [77]. However, we found that the effect of YAP1 on patient survival was unclear in the entire breast cancer population from public databases. A recent study showed that YAP1 suppressed ER+ breast cancer by inhibiting the expression of ESR1 via VGLL3 [35]. We explored the effect of YAP1 on survival in different molecular subtypes of breast cancer (ER+, HER2+, ER−HER2−) from cBioPortal, cell proliferation assays in vitro and our tissue microarrays. We found that high YAP1 expression was associated with longer OS in patients with ER+ breast cancer, whereas no association was found in patients with other subtypes from cBioPortal. In an in vitro environment, YAP1 inhibited the cell proliferation of three breast cancer cell subtypes (MCF7 cells belong to ER+, SKBR3 cells belong to HER2+, MDA-MB-231 cells belong to ER−HER2−). In tissue microarrays, total YAP1 was associated with shorter OS in breast cancer patients. Measuring breast cancer patient survival with total YAP1 may not be the best approach. Furthermore, we found that overexpressed YAP1 was mainly present in the cytoplasm and that cytoplasmic mutant YAP1S127D inhibited the proliferation of breast cancer cells, which was inconsistent with previous studies showing that YAP1 promoted tumor cell proliferation [3, 4]. This functional inconsistency was of great interest to us. Analysis of the literature suggested that the pro-proliferation effect of YAP1 observed in previous studies mainly depended on its identity as a transcriptional co-activator in the nucleus. YAP1 is not a classical nuclear transcription factor; it binds with members of the TEAD transcription factor family, promoting DNA binding of those transcription factors and thus transcription of downstream target genes [76, 78]. Therefore, we investigated the association between YAP1 target genes CTGF and CYR61 (which represent the nuclear activity of YAP1 to some extent) and the survival of breast cancer patients using public databases. Simultaneous high expression of CTGF and CYR61 was not associated with the survival of breast cancer patients, but might be caused by the fact that the expression of CTGF and CYR61 could not fully represent the nuclear activity of YAP1. Although YAP1 was known to interact with the WNT pathway [79, 80] and to be easily degraded in the cytoplasm [69, 81, 82], studies regarding the role of YAP1 in the cytoplasm are very few and have produced unclear results. YAP1 can shuttle between the nucleus and the cytoplasm, but was mainly expressed in the cytoplasm in breast cancer tissues. This suggests that cytoplasmic YAP1 may exert an opposite effect on breast cancer cell proliferation than nuclear YAP1. This hypothesis was confirmed in the present study through in vitro cell culture experiments, human breast cancer tissue microarrays, and in vivo nude mouse xenograft experiments.
Autophagy is an evolutionarily conserved intracellular catabolic degradation process. Currently, the bidirectional role of autophagy in tumors is controversial. Regarding the direction of autophagy to promote or suppress cancer, there are two viewpoints: “Degree Theory” and “Microenvironment Theory”. The former believes that autophagy is a form of programmed cell death, and the continuous activation of autophagy will lead to cell degradation of necessary components towards death [14]. A certain degree of autophagy has a protective effect on tumor cells and can promote tumor progression and drug resistance [83, 84]. The latter is believed to be related to the microenvironment, that is, in the early stage of tumorigenesis, autophagy as a quality control mechanism, it inhibits tumorigenesis and early tumor progression [85]. Once the tumor develops to an advanced stage and is under environmental stress, it can act as a survival mechanism to promote tumor growth and malignant phenotype [86, 87].
We found that cytoplasmic YAP1 increased the basal level of autophagy in breast cancer cells. In addition, NLS-YAP15SA (promotes the nuclear localization of YAP1 and increases YAP1 transcriptional activity) also induced autophagy, consistent with published research [16, 88], but YAP1S127D had the most significant effect. YAP1 promoted autophagy in breast cancer cells mainly in the cytoplasm. Using treatment with the autophagy inhibitor CQ and mRFP-GFP-LC3, we observed that YAP1 promoted the formation of AP and that the flow of autophagy was smooth.
Currently, the concept that cell death is completely dependent on autophagy is controversial. There are three types of autophagic cell death: autophagy-associated cell death (autophagy simply accompanies the cell death process and does not have an active role in it), autophagy-mediated cell death (autophagy induction triggers apoptosis), and autophagy-dependent cell death (a distinct mechanism of cell death that occurs independently of apoptosis or necrosis) [89-91]. In the present study, we demonstrated that the induction of apoptosis was suppressed by inhibiting autophagy, indicating that YAP1 caused autophagy-mediated cell death.
Previous research on the formation of AP had confirmed the involvement of the mTOR-ULK1 complex, autophagy-related protein ATGs, the coat protein complex II (COPII), and the ESCRT mechanism, among other components [21, 23, 92]. The membrane cleavage function of ESCRTs played a key role in the closure of bowl-shaped bilayer vesicles to form the AP [23, 92-96]. Our research group used bioinformatics and molecular docking methods to determine that YAP1 might promote AP formation through ESCRTs. qRT-PCR confirmed that YAP1 could not promote the transcription of ESCRT-III subunits, and co-IP showed that YAP1 binding to the ESCRT-III subunits CHMP2B and VPS4B promoted assembly of the CHMP4B-VPS4B complex. Furthermore, co-localization of YAP1 with CHMP2B and VPS4B in the cytoplasm was observed via IF staining. This function of YAP1 in promoting autophagic cell death was carried out in the cytoplasm, independent of classical transcriptional co-activation in the nucleus.
In order to clarify that autophagic cell death was mediated by cytoplasmic YAP1, we treated cells with EGCG; this treatment did not change the level of total YAP1, but localized YAP1 to the cytoplasm by activating the Hippo pathway, as confirmed in previous studies [97, 98]. Through in vitro and in vivo experiments, we found that EGCG could promote assembly of the ESCRT-III complex and subsequent autophagic death of breast cancer cells, and inhibited xenograft growth by increasing retention of phosphorylated YAP1 in the cytoplasm. Finally, we used human breast cancer tissue microarrays to analyze cytoplasmic YAP1 expression and found that high cytoplasmic YAP1 expression was associated with longer survival, validating our hypothesis.
The role of YAP1 in tumors mainly depends on its abundance and subcellular localization [99, 100]. Protein ubiquitination is an important way to maintain protein abundance. E3 ubiquitin ligases determine the specificity of ubiquitinated substrates [101]. In contrast, deubiquitinase dissociates ubiquitin molecules from ubiquitinated proteins, inhibiting degradation and increasing protein abundance [102]. We hypothesized that cytoplasmic YAP1 abundance in breast cancer cells was maintained by the ubiquitin-proteasome system, and verified with mass spectrometry that the ubiquitin ligase NEDD4L promoted ubiquitin-mediated degradation of YAP1. IHC staining and WB analysis confirmed that NEDD4L was negatively correlated with YAP1 expression in human breast cancer tissues, suggesting that interference with NEDD4L expression could be used to maintain cytoplasmic YAP1 abundance.
Although the present study constructed a cytoplasmic mutant of YAP1 and used EGCG as a tool to determine the effect of cytoplasmic YAP1 on breast cancer in vitro, only EGCG was used as a model in vivo. The number of breast cancer tissue microarray samples was small and might produce false positives. Our next focus will be to use the breast cancer cell line stably transfected with YAP1S127D to carry out nude mouse xenograft experiments, or use tissue-specific expression of YAP1S127D transgenic mice to simulate the in vivo environment to further study the effect of cytoplasmic YAP1 on breast cancer, and continue to expand clinical sample sizes to make research more relevant. The functions of YAP1 in inhibiting cell proliferation and promoting apoptosis were partially attenuated but not completely reversed by CQ and siATG7, indicating that there may be other mechanisms for YAP1 to inhibit breast cancer. It seems that the inhibition of P73 expression by nuclear YAP1 in the literature may provide a partial explanation [9, 10]. The impact of YAP1 on breast cancer fate still needs to be further explored.
5 CONCLUSIONS
In conclusion, our experimental results demonstrated that cytoplasmic YAP1 mediated the assembly of the ESCRT-III complex subunits CHMP2B and VPS4B to promote autophagic death of breast cancer cells. Furthermore, the ubiquitin ligase NEDD4L mediated ubiquitinated degradation of cytoplasmic YAP1. The results of the present study clarified the role of YAP1 in breast cancer and provided new avenues for breast cancer treatments through targeting YAP1.
DECLARATIONS
AUTHOR CONTRIBUTIONS
Xuesong Chen: Contributed to conception design and the supervision of this project. Yan Guo, Yuqing Cui, and Yangyang Li: Conducted most of the experiments including cell biology, molecular biology and nude mouse xenograft experiments, and drafted the manuscript. Xiaoying Jin, Dandan Wang, and Mengxia Lei: Analyzed and interpreted of the data. Fengzhi Chen ang Yali Liu: Contributed to the download and analysis of data from the public databases. Jinwen Xu, Guanyu Yao, and Guangchun Zeng: Contributed to the collection and analysis of clinical samples. All authors read and approved the final manuscript.
ACKNOWLEDGEMENT
We thank the Heilongjiang Institute of Cancer Prevention and Treatment, the Biotherapy Center of Harbin Medical University Cancer Hospital and the main campus of Harbin Medical University for providing the experimental platforms.
CONFLICT OF INTERESTS STATEMENT
The authors declare no conflict of interest.
CONSENT FOR PUBLICATION
Not applicable.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Breast cancer tissues were taken from patients undergoing surgical resection at the Harbin Medical University Cancer Hospital (Harbin, China). All experiments using these tissues were approved by the Ethics Committee of Harbin Medical University (Ethical approval number KY2016-13, KY2018-09, KY2022-27), and written informed consent was obtained from all patients. Animal experiments were performed according to the guidelines recommended by the Institutional Animal Care and Use Committee (IACUC) of Harbin medical university (GB/T 35892-2018) and the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals.
Open Research
DATA AVAILABILITY STATEMENT
The survival datasets used in the present study are available from The Cancer Genome Atlas (TCGA) database (https://tcga-data.nci.nih.gov/), cBioPortal database (https://www.cbioportal.org/), and the Gene Ommnibus (GEO) database (http://ncbi.nlm.nih.gov/geo/,GSE56245). The protein structures used for molecular docking were downloaded from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (https://www.rcsb.org/) and the AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk/). The online databases and programs used were Natural Product Activity & Species Source (NPASS) database (http://bidd.group/NPASS/), Kaplan-Meier (https://kmplot.com/analysis/), STRING (https://cn.string-db.org), UbiBrowser (http://ubibrowser.ncpsb), STITCH (http://stitch.embl.de/), DAVID (https://david.ncifcrf.gov/tools.jsp), and ZDOCK SERVER (https://zdock.umassmed.edu/). All other data are available from the corresponding author upon reasonable request.