Novel endogenous endoplasmic reticulum transmembrane protein SURF4 suppresses cell death by negatively regulating the STING-STAT6 axis in myeloid leukemia
Abbreviations
-
- ROS
-
- Reactive oxygen species
-
- ER
-
- endoplasmic reticulum
-
- STING
-
- stimulator of interferon genes
-
- STAT6
-
- signal transducer and activator of transcription 6
-
- SURF4
-
- surfeit 4
-
- TBK1
-
- TANK-binding kinase 1
-
- shRNA
-
- short hairpin RNA
-
- sgRNA
-
- single-guide RNA
-
- AML
-
- acute myeloid leukemia
-
- scRNA-seq
-
- single-cell RNA sequencing
-
- UMAP
-
- uniform manifold approximation and projection
-
- HSCs
-
- hematopoietic stem cells
-
- cDCs
-
- conventional dendritic cells
-
- GO
-
- Gene Ontology
Myeloid differentiation was shown to be associated with reduced leukemic cell burden and leukemia-initiating cells and improved survival [1]. Reactive oxygen species (ROS) are associated with leukemia and can induce endoplasmic reticulum (ER) stress. ER stress induces several mechanisms, including cell death [2]. The stimulator of interferon genes (STING) is also an ER transmembrane protein and promotes anti-tumor immunity by linking innate and adaptive immunity [3]. Signal transducer and activator of transcription 6 (STAT6) plays a prominent role in adaptive immunity by transducing signals from extracellular cytokines and inducing apoptosis in cancer cells [4]. Surfeit 4 (SURF4) is a multi-pass ER transmembrane protein that participates in the ER-Golgi compartment, and SURF4 was found to be amplified and highly expressed in leukemic cells (Supplementary Figure S1A) and several cancer types, including blood cancers [5]. STING interacts with STAT6 in the ER, and TANK-binding kinase 1 (TBK1) activates pSTAT6Y641, leading to anti-tumor effects in cancer cells [6]. Interestingly, SURF4 binds to STING [7], but it remains unclear how the STING-TBK1-STAT6 axis yields anti-tumor effects in blood cancers. The study methods are detailed in the Supplementary Materials.
To determine the function of SURF4 in myeloid leukemic cells, we transduced multiple short hairpin RNA (shRNA) constructs targeting SURF4 in myeloid malignancies. Three shRNAs targeting distinctive regions within the SURF4 transcript provided effective knockdown in THP1, HL60 and K562 myeloid leukemic cells (Supplementary Figure S1B). SURF4 shRNA-transfected THP1, HL60 and K562 myeloid leukemic cells showed significantly reduced proliferation compared to control cells (Figure 1A). Further, we found an increased number of apoptotic cells in SURF4 shRNA-mediated THP1, HL60 and K562 myeloid leukemic cells (Figure 1B, Supplementary Figure S1C) and single-guide RNA (sgRNA) deletion of Surf4 in normal hematopoietic progenitor cells (Supplementary Figure S1D). It is reported that AKT and ERK signaling pathways are implicated in myeloid leukemia, ROSs are associated with leukemia, and prolonged ROS elevation to activate the Jun N-terminal kinase (JNK)/c-JUN signaling pathway [8]. Increased phospho-JNK (pJNK) expression and decreased pERK and pAKT expression were detected in these cells (Supplementary Figures S1E-F). Interestingly, SURF4 shRNA-mediated THP1 and HL60 cells and sgRNA deletion of Surf4 in Mll/Af9 cells exhibited significantly increased differentiation (Supplementary Figure S1G-H) and accumulation of ROS (Supplementary Figure S1I), respectively. In addition, the silencing of SURF4 led to tumor growth arrest in vivo (Figure 1C). No differences in the cell cycle status were observed from the SURF4 shRNA-mediated THP1 and K562 cells (Supplementary Figure S1J). Collectively, SURF4 suppressed myeloid differentiation and ROS production and inhibited cell death in myeloid leukemic cells.
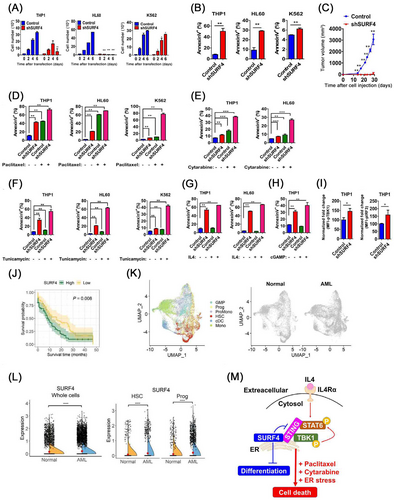
Impact of SURF4 on myeloid leukemic cells. (A) SURF4 shRNA-mediated THP1, HL60, and K562 cells were counted every two days. (B) Quantification of apoptotic cells from SURF4 shRNA-mediated THP1, HL60, and K562 cells. (C) Tumorigenicity was analyzed with a xenograft model of NOD-SCID mice. Subcutaneous injection of SURF4 shRNA-mediated HL60 cells was performed and tumor growth was quantified (n = 5). (D-F) Quantification of the apoptotic cells from SURF4 shRNA-mediated THP1, HL60, and K562 cells after paclitaxel (D), cytarabine (E), or tunicamycin (F) treatment. (G) Quantification of the apoptotic cells from SURF4 shRNA-mediated THP1 and HL60 cells after IL4 treatment. (H) Quantification of the apoptotic cells from SURF4 shRNA-mediated THP1 cells after cGAMP treatment. (I) Quantification of the normalized fold change in MFI for the indicated phospho-protein in SURF4 shRNA-mediated THP1 cells. (J) Survival analyses for SURF4. Overall survival for 4 years with Kaplan-Meier curve stratified by SURF4 expression. (K) UMAP visualization of cells from normal and AML patients. In the UMAP plot, each point represents a cell, and the space of the point reflects the location of the cell in low-dimensional space based on transcriptional similarity. The left panel is colored by the cell type of bone marrow, and the right panel shows UMAPs separated by donor origin. (L) Gene expression levels of SURF4 in whole cells, progenitors, and HSCs. The Y-axis of the violin plot is log-normalized counts of SURF4, and the described p-value was derived using the Wilcox rank-sum test comparing the gene expression levels in the normal and AML cells. (M) SURF4 negatively regulates STAT6 and STING functions and suppresses differentiation and cell death in myeloid leukemic cells. Error bars indicated the S.E.M. (**** P ≤ 0.001, *** P ≤ 0.001, ** P ≤ 0.01, * P ≤ 0.05). (n = 2 independent experiments and 3 total measurements per group and treatment).
Abbreviations: ROS, Reactive oxygen species; ER, endoplasmic reticulum; STING, stimulator of interferon genes; STAT6, signal transducer and activator of transcription 6; SURF4, surfeit 4; sgRNA, single-guide RNA; AML, acute myeloid leukemia; scRNA-seq, single-cell RNA sequencing; UMAP, uniform manifold approximation and projection; cDCs, conventional dendritic cells; GO, Gene Ontology.
Prolonged ER stress initiates the mechanisms of cell death [2]. Signaling apoptosis in response to ER stress is mainly associated with the apoptotic PKR-like ER kinase (PERK), which phosphorylates the eukaryotic initiation factor 2α (eIF2α) and C/EBP homologous protein (CHOP) axis. We then tested whether SURF4 contributed to the cell death effects of combinatorial treatments with paclitaxel (Figure 1D, Supplementary Figure S2A), cytarabine (Figure 1E, Supplementary Figure S2B), tunicamycin (Figure 1F, Supplementary Figure S3A), interleukin 4 (IL4) (Figure 1G, Supplementary Figure S3B), or STING agonist cyclic GMP-AMPs (cGAMP) (Figure 1H, Supplementary Figure S3C) on SURF4 shRNA-mediated myeloid leukemic cells, respectively. These molecules were shown to activate ER stress, STAT6 and STING signaling pathways, and led to cell death in cancer cells [2-4, 6]. Increased cell death was detected in SURF4 shRNA-mediated myeloid leukemic cells after combinatorial treatment with paclitaxel, cytarabine, tunicamycin, IL4, or cGAMP. Western blotting after paclitaxel, tunicamycin, IL4, or cGAMP treatment of SURF4 shRNA-mediated THP1 cells showed activation of cleaved caspase 9, caspase 3, cleavage of poly(ADP-ribose) polymerase 1 (PARP1) (Supplementary Figure S4A), phospho-eukaryotic initiation factor 2α (peIF2α), activating transcription factor 4 (ATF4), C/EBP homologous protein (CHOP) (Supplementary Figure S4B), pSTAT6 (Supplementary Figure S4C-D), pTBK1 and pIRF3 (Supplementary Figure S4E), respectively. We quantified the intensity of these western blots. Silencing of SURF4 increased pSTAT6 levels in association with depleted SURF4 levels with and/or without IL4 treatment (Supplementary Figure S4D, F). pTBK1 and pIRF3 are downstream targets of the STING signaling pathway, and increased pTBK1 and pIRF3 expression were detected in SURF4 shRNA-mediated THP1 cells (Figure 1I, Supplementary Figure S4E). Collectively, depletion of SURF4 synergistically induces apoptosis in myeloid leukemic cells via anti-cancer drugs, such as paclitaxel, cytarabine, and ER stress inducers, such as tunicamycin. IL4-dependent pSTAT6 and/or STING activation-induced apoptosis was also increased in SURF4-silenced leukemic cells.
Next, we explored the overall survival of acute myeloid leukemia (AML) patients based on relative levels of SURF4 expression from the cancer genome atlas (TCGA). Individuals with high SURF4 expression had significantly shorter survival than those with low SURF4 expression (Figure 1J). Further, to characterize the expression of SURF4 in AML, we downloaded and explored the AML single-cell RNA sequencing (scRNA-seq) datasets [9] (Figure 1K). The scRNA-seq data from bone marrow (BM) cells of normal healthy donors and AML patients were subjected to uniform manifold approximation and projection (UMAP) analysis. SURF4 was highly expressed in the total population and displayed remarkably high expression in hematopoietic stem cells (HSCs) and progenitor cells from AML patients (Figure 1L). However, there are no differences in SURF4 expression in the AML cells from conventional dendritic cells (cDCs), granulocyte-macrophage progenitor (GMP) cells, monocytes (mono), and promonocytes (promono) compared with normal healthy donors (Supplementary Figure S5A). Gene ontology (GO) analysis revealed that SURF4 might be associated with differentially expressed immune responses [3, 4, 6, 7] (Supplementary Figure S5B). Collectively, the expression of SURF4 was significantly increased in AML patients, suggesting that SURF4 is relevant to the pathogenesis of hematological malignancies (Figure 1M).
In this study, we demonstrated that SURF4 suppressed myeloid differentiation and inhibited cell death in myeloid leukemic cells via negatively regulating the STING-TBK1-STAT6 axis. Thus, we propose that the inhibition of SURF4, such as using monoclonal antibodies and/or aptamer, may be used as a potential therapeutic strategy for the treatment of hematological malignancies.
DECLARATIONS
AUTHOR CONTRIBUTIONS
J.K. and H.L. designed the research, analyzed the data, and wrote the manuscript. J.K. carried out most of the experimental work with the help of H.L., C.M.H., J.H.N., H.J.Y., W.H.C., H.S.K., and C.H. Y.H.K. and D.L. directed the research.
CONFLICT OF INTEREST
No conflicts of interest to disclose.
FUNDING
This work was supported by grants from the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (2021R1A2C4001466 and 2022R1A5A2027161 for D.L. and 2020R1C1C1003741 and 2018R1A5A2023879 for Y.H.K).
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The animal experiments were approved by the Animal Ethics Committee of the Pusan National University School of Medicine (PNU-2022-0141).
CONSENT FOR PUBLICATION
Not applicable.
Open Research
DATA AVAILABILITY STATEMENT
The scRNA-seq bone marrow cell data from normal and AML patients were downloaded from the GSE116256 database. The data used and/or analyzed for this study will be available from the corresponding author upon reasonable request.




