PDGFA-associated protein 1 is a novel target of c-Myc and contributes to colorectal cancer initiation and progression
Abstract
Background
The mechanism underlying colorectal cancer (CRC) initiation and progression remains elusive, and overall survival is far from satisfactory. Previous studies have shown that PDGFA-associated protein 1 (PDAP1) is upregulated in several cancers including CRC. Here, we aimed to identify the cause and consequence of PDAP1 dysregulation in CRC and evaluate its role as a potential therapeutic target.
Methods
Multi-omics data analysis was performed to identify potential key players in CRC initiation and progression. Immunohistochemistry (IHC) staining was applied to determine the expression pattern of PDAP1 in CRC tissues. Pdap1 conditional knockout mice were used to establish colitis and CRC mouse models. RNA sequencing, a phosphoprotein antibody array, western blotting, histological analysis, 5-bromo-2′-deoxyuridine (BrdU) incorporation assay, and interactome analysis were applied to identify the underlying mechanisms of PDAP1. A human patient-derived xenograft (PDX) model was used to assess the potential of PDAP1 as a therapeutic target.
Results
PDAP1 was identified as a potential key player in CRC development using multi-omics data analysis. PDAP1 was overexpressed in CRC cells and correlated with reduced overall survival. Further investigation showed that PDAP1 was critical for the regulation of cell proliferation, migration, invasion, and metastasis. Significantly, depletion of Pdap1 in intestinal epithelial cells impaired mucosal restitution in dextran sulfate sodium salt-induced colitis and inhibited tumor initiation and growth in colitis-associated cancers. Mechanistic studies showed that c-Myc directly transactivated PDAP1, which contributed to the high PDAP1 expression in CRC cells. PDAP1 interacted with the juxtamembrane domain of epidermal growth factor receptor (EGFR) and facilitated EGFR-mitogen-activated protein kinase (MAPK) signaling activation, which resulted in FOS-related antigen 1 (FRA-1) expression, thereby facilitating CRC progression. Notably, silencing of PDAP1 could hinder the growth of patient-derived xenografts that sustain high PDAP1 levels.
Conclusions
PDAP1 facilitates mucosal restitution and carcinogenesis in colitis-associated cancer. c-Myc-driven upregulation of PDAP1 promotes proliferation, migration, invasion, and metastasis of CRC cells via the EGFR-MAPK-FRA-1 signaling axis. These findings indicated that PDAP1 inhibition is warranted for CRC patients with PDAP1 overexpression.
Abbreviations
-
- AOM
-
- azoxymethane
-
- AP-1
-
- activator protein-1
-
- bp
-
- base pairs
-
- BrdU
-
- 5-bromo-2′-deoxyuridine
-
- BSA
-
- bovine serum albumin
-
- CAC
-
- colitis-associated cancer
-
- CAN
-
- copy number alteration
-
- ChIP
-
- chromatin immunoprecipitation
-
- CRC
-
- colorectal cancer
-
- DAPI
-
- 4′,6-diamidino-2-phenylindole
-
- DEG
-
- differentially expressed gene
-
- DSS
-
- dextran sulfate sodium
-
- ECD
-
- extracellular domain
-
- EGFR
-
- epidermal growth factor receptor
-
- FDR
-
- false discovery rate
-
- FITC
-
- fluorescein isothiocyanate
-
- FRA-1
-
- FOS-related antigen 1
-
- GEO
-
- gene expression omnibus
-
- GSEA
-
- gene set enrichment analysis
-
- HE
-
- hematoxylin and eosin stain
-
- IHC
-
- immunohistochemistry
-
- IPTG
-
- isopropylthio-β-galactoside
-
- IP
-
- immunoprecipitation
-
- JM
-
- juxtamembrane
-
- KEGG
-
- Kyoto Encyclopedia of Genes and Genomes
-
- loxP
-
- locus of x-over of phage P1
-
- Luc
-
- luciferase
-
- MAPK
-
- mitogen-activated protein kinase
-
- MEK1
-
- MAPK/ERK kinase 1
-
- NES
-
- normalized enrichment score
-
- OS
-
- overall survival
-
- PCNA
-
- proliferating cell nuclear antigen
-
- PDAP1
-
- PDGFA-associated protein 1
-
- PDGF
-
- platelet-derived growth factor
-
- PDX
-
- patient-derived xenograft
-
- PKB
-
- protein kinase B
-
- shCtrl
-
- scramble control of PDAP1 knockdown
-
- shMYC
-
- knockdown of MYC
-
- shPDAP1
-
- knockdown of PDAP1
-
- siPDAP1
-
- siRNAs targeting PDAP1
-
- TCGA
-
- the cancer genome atlas
-
- TM
-
- transmembrane domain
-
- TUNEL
-
- terminal deoxynucleotidyl transferase dUTP nick-end labeling
-
- RFP
-
- red fluorescent protein
-
- qPCR
-
- quantitative polymerase chain reaction
1 BACKGROUND
Colorectal cancer (CRC) is the second most deadly cancer worldwide, accounting for approximately 10% of all annually diagnosed cancers and cancer-related deaths [1, 2]. More than 90% of CRCs contain a mutation in APC or CTNNB1. Although colitis-associated cancer (CAC) has a lower frequency of somatic APC mutation compared to sporadic CRC [3-6], Wnt pathway activation and nuclear accumulation of β-catenin are prevalent in CAC [7, 8]. The stabilized β-catenin leads to deregulated expression of downstream target genes, including the c-Myc proto-oncogene [9]. Intercepting and blocking the c-Myc pathway at various points in the signaling cascade is an attractive approach for CRC chemoprevention and therapeutics [10].
PDAP1, also known as the 28 kDa heat- and acid-stable phosphoprotein, is highly conserved across vertebrate taxa. It was originally characterized as a novel casein kinase II substrate in the rat brain [11] and was associated with platelet-derived growth factor (PDGF)-A with low affinity in a rat neural retina cell line [12]. Most recently, PDAP1 has been identified as an essential regulator of cellular homeostasis in mature B cells by protecting them from stress-induced cell death and promoting antibody gene diversification [13]. PDAP1 has been found to be overexpressed in gastric cancer [14], rectal carcinoma [15] and grade IV glioma, and to promote the proliferation of malignant glioma cells [16]. However, the causes and consequences of PDAP1 dysregulation in cancer are poorly understood. Single-cell genomic techniques have become useful for segregating specific processes in the intricate ecosystems of human tumors [17], so we used these techniques to identify the role of PDGFA-associated protein 1 (PDAP1) dysregulation in CRC.
In this study, we evaluated the expression status of PDAP1 in CRC tissues and paired paracancerous tissues, and the prognostic value of PDAP1 in CRC. Then in vivo and in vitro models were used to analyze the functions of PDAP1 in CRC development and its underlying mechanisms of action. We also evaluated the potential of PDAP1 as a therapeutic target in a human patient-derived xenograft (PDX) model. We anticipated that targeting PDAP1 may be a potential therapeutic strategy for CRC.
2 MATERIALS AND METHODS
2.1 Human tissues
Surgically resected CRC tissues were collected from the Department of Gastrointestinal Surgery, Xijing Hospital, which is affiliated with the Fourth Military Medical University (Xi'an, Shaanxi, China). Patients with more than one primary tumor were excluded from the study. All eligible patients were diagnosed between December 1, 2010, and April 25, 2017. All the cases had been followed up annually and the last date of follow-up was July 2019. The staging classification was based on the tumor, node, metastasis (TNM) staging by AJCC, 8th edition. The resected tissue was divided into two parts. One part was stored at −80°C, and the other part was formalin-fixed and paraffin-embedded. Experienced pathologists from the Xijing Hospital reviewed the tissue sections. All participants provided written informed consent, and the study was approved by the Hospital Ethics Committee (KY20213194-1). CRC tissue arrays were obtained from Outdo Biotech (Shanghai, China).
2.2 Mice
Male mice with a C57BL/6 background weighing approximately 22 g were used in this study. Pdap1fl/fl mice were generated by Biocytogen (Beijing, China). As shown in Supplementary Figure S1A, the loxP fragments were inserted into introns 2 and 4. Exons 3 and 4 were conditionally removed using Cre recombinase. Southern blot analysis was performed during F1 mouse screening to exclude the possibility of random integration (Supplementary Figure S1B-C). Villin-Cre mice were obtained from Jackson Laboratory (Bar Harbor, ME, USA). Immunodeficient nude mice (BALB/c, 6-8 weeks old) were obtained from Beijing HFK Bioscience (Beijing, China). All animal protocols were approved by the Animal Care and Welfare Committee of the Fourth Military Medical University. All the mice used in this study were housed in a specific pathogen-free facility.
2.3 Live cell microscopy
Live cell fluorescence imaging was performed as described previously [18]. Briefly, the reporter plasmid pCCC-TagRFP (PCNA-cb, Chromotek, Planegg-Martinsried, Germany) was used to obtain stable cell lines via random plasmid integration. A total of 5000 cells were seeded in 96-well imaging plates (Corning, NY, USA). Images were acquired at 37°C at 35 min intervals using Cytation 5 (BioTek Instruments, Winooski, VT, USA) 20 × objective and analyzed using Gen5 (Agilent, Palo Alto, CA, USA) and ImageJ (National Institutes of Health, Bethesda, MD, USA). Median nuclear red fluorescent protein (RFP) intensity was used as a surrogate measurement for DNA replication.
2.4 Dextran sulfate sodium (DSS)-induced colitis
Male mice, weighing approximately 22 g, were used in this study. A 2.5% solution of DSS (MP Biomedicals, Santa Ana, CA, USA) in drinking water was administered for the first five days, followed by regular drinking water for another five days. The mice were monitored daily for body weight. On day 10, they were euthanized, and colon lengths were measured.
2.5 Azoxymethane (AOM)/DSS-induced CAC
Male mice, weighing approximately 22 g, were used in this study. They were treated with an intraperitoneal injection of a single dose of AOM (12.5 mg/kg, MP Biomedicals). After three days, a 2% solution of DSS in drinking water was administered for five days, followed by regular drinking water for another two weeks. The DSS treatment was repeated for two additional cycles. On day 120, the mice were euthanized and the colon tumors were dissected for further analysis.
2.6 Analysis of intestinal permeability and BrdU incorporation
Fluorescein isothiocyanate (FITC)-conjugated dextran with an average molecular weight of 40000 (Merck, Kenilworth, NJ, USA) was dissolved at a concentration of 100 mg/mL in PBS. BrdU (Beyotime, Shanghai, China) was dissolved at a concentration of 20 mg/mL in PBS. FITC was administered to each mouse (500 mg/kg) via oral gavage. One hour later, the mice were intraperitoneally injected with BrdU (100 mg/kg). After 3 h, mice were anesthetized, and 300 μL of blood per mouse was collected. The concentration of FITC-dextran was determined using Cytation 5 with a GFP filter cube. Serum from untreated mice was used to determine the background.
2.7 His pull-down and mass spectrometry
The precooled CRC tissues were ground using a TissueLyser II (QIAGEN, Venlo, Netherlands) at 28 Hz for 5 min and then lysed with lysis buffer. His pull-down assays were performed using Pierce™ His Protein Interaction Pull-Down kit (Pierce, Rockford, IL, USA), where His6-tagged PDAP1 proteins were the bait, and the lysates from the fresh CRC tissues described above were the prey. The eluted proteins were identified by mass spectrometry. An LTQ-Orbitrap hybrid mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) was used to obtain tandem mass spectra of the tryptic peptides. The resulting MS/MS spectra were analyzed using a Thermo Scientific Proteome Discoverer (version 2.5; Thermo Fisher Scientific) for putative protein identification.
2.8 Chromatin immunoprecipitation (ChIP) assay
The binding of c-Myc to the PDAP1 promoter was measured using a ChIP assay as previously described [19]. HCT-8 cells were fixed with 1% formaldehyde and treated with 1 mol/L glycine. After washing with ice-cold PBS, cells were scraped off and lysed with 1% SDS lysis buffer supplemented with a protease inhibitor cocktail on ice. The lysed cells were sonicated to shear the chromatin DNA to an optimal size of approximately 200 bp-1000 bp. Sheared chromatin DNA was immunoprecipitated using antibodies against c-Myc (Santa Cruz Biotechnology, Dallas, TX, USA). Equal amounts of IgG (Santa Cruz Biotechnology) served as negative controls. The immunoprecipitate was then incubated with protein A/G magnetic beads (Millipore, Boston, MA, USA) and pulled down on a magnetic stand. After washing, reverse crosslinking was performed in 5 mol/L NaCl at 65°C overnight. Contaminating RNA was cleaned with ribonuclease A, and the protein was digested with proteinase K. Finally, the sheared DNA recovered from reverse crosslinking was extracted using a DNA extraction kit for further analysis with quantitative polymerase chain reaction (qPCR). The same amount of sheared DNA without antibody precipitation after reverse crosslinking was used as the input control.
2.9 Clonogenic assay
Clonogenic assays were performed as previously described [20]. Briefly, cells were collected and inoculated into 6-well plates. The cells were cultured for 2 weeks until visible clones emerged and fresh medium was replaced every 4 days. After being washed twice with PBS, the cells were fixed and stained with 0.2% crystal violet (Beyotime) in 95% ethanol at 25°C for 30 min. The plates were washed several times with PBS to remove excess dye. The cell clones in each well were counted.
2.10 RNA sequencing
RNA sequencing was performed as previously described [21]. Sequence libraries were generated and sequenced by CapitalBio Technology (Beijing, China). The clean reads were aligned to the reference genome and the processed reads from each sample were aligned using HISAT. Cuffdiff was used to analyze the differentially expressed genes between the samples.
2.11 PDX mouse model
PDX mouse models were obtained from BEIJING IDMO Co. (Beijing, China). For expansion, 2 × 106 cells in 0.1 mL RPMI 1640 medium were subcutaneously inoculated into the right posterior flank of nude mice. Two weeks after tumor cell implantation, mice were randomly allocated and administered in vivo-optimized small interfering RNAs (siRNAs). After four rounds of treatment, the mice were euthanized and the tumors were removed for analysis.
2.12 Statistical analysis
Quantitative results are presented as the mean ± standard deviation, and individual data points are plotted. All statistical analyses were performed for each panel. Differences were compared using two-tailed Student's t-test, as indicated in the figure legends. Spearman's R was used to determine the correlation between the relative expression levels of the genes or proteins. All reported P-values were two-tailed, and P < 0.05 was considered significant.
Detailed methods are available in the Supplemental materials.
3 RESULTS
3.1 Augmented PDAP1 in CRC correlates with poor survival
To identify the potential key players in CRC initiation and progression, we reanalyzed the single-cell RNA sequencing data of 63689 cells from 23 primary CRC and 10 matched normal mucosa samples (GSE132465) [22], and the proteome of 146 CRC tissues with paired remote normal tissues [23]. Of the 2307 differentially expressed genes (DEGs) with a fold change ≥ 1.5 and adjusted P < 0.05 between 17469 tumor cells and 1070 normal epithelial cells (Supplementary Figure S2A), 104 DEGs exhibiting highly similar expression patterns at the proteomic level were selected for further investigation of their effects on overall survival (OS) using the Gene Expression Omnibus (GEO) database (Supplementary Figure S2B). Of the 54 genes with significant effects on OS (P < 0.01), PDAP1 was brought to our attention because its regulation is obscured by bulk genomic analysis using The Cancer Genome Atlas (TCGA) data and its biological function remains almost unknown, especially in cancer (Supplementary Figure S2C). Immunohistochemistry was performed to examine PDAP1 expression in two independent cohorts of CRC biopsies (Supplementary Table S1 and Supplementary Table S2). The positive expression rates of PDAP1 in CRC tissues were 83.57% (234/280) and 96.00% (72/75), respectively. In paired paracancerous tissues, the positive expression rate of PDAP1 were 23.93% (67/280) and 25.33% (19/75), respectively. The expression of PDAP1 in cancer tissues was significantly higher than that in cancer-adjacent tissues (Supplementary Figure S3A-B). Furthermore, we determined the effects of PDAP1 expression on the prognosis of patients with CRC. We found that the survival of patients with high PDAP1 expression was worse than that of patients with low PDAP1 expression (Supplementary Figure S3C).
3.2 PDAP1 promotes CRC progression
To determine the role of PDAP1 in CRC progression, we evaluated PDAP1 expression in several CRC cell lines (Supplementary Figure S4A) and established stable PDAP1 knockdown (shPDAP1) cell lines (Supplementary Figure S4B-C). The clonogenic assay showed that the number of colonies derived from PDAP1 knockdown cells was significantly reduced (Supplementary Figure S4D). We observed that PDAP1 knockdown decreased cell migration (Supplementary Figure S4E), invasion (Supplementary Figure S4F), and proliferation (Supplementary Figure S4G). Cell cycle analysis showed that PDAP1 knockdown arrested the cell division cycle during the S/G2 phase transition (Supplementary Figure S4H). To monitor DNA replication in living cells, cells were transfected with a marker for DNA replication (proliferating cell nuclear antigen [PCNA]-cb) (Supplementary Figure S4I). We found that PDAP1 knockdown cells displayed weak and prolonged PCNA-cb foci compared to control cells (Supplementary Figure S4J), indicating that completion of DNA replication was delayed by PDAP1 knockdown. In sharp contrast, PDAP1 knockdown had little effect on sphere formation (Supplementary Figure S5A) and apoptosis (Supplementary Figure S5B). Furthermore, cells overexpressing PDAP1 showed enhanced colony formation (Supplementary Figure S5C), migration (Supplementary Figure S5D), invasion (Supplementary Figure S5E), and proliferation (Supplementary Figure S5F).
To test the tumorigenic potential of PDAP1 in vivo, we performed a xenograft assay using immunodeficient mice. Tumor progression was significantly decreased in mice injected with PDAP1 knockdown cells (Supplementary Figure S5G-I). Furthermore, we used a tail vein injection assay to investigate whether the metastatic ability of tumor cells was influenced by PDAP1 in vivo. Sixty days after injection, PDAP1 knockdown cells formed significantly less metastatic nodules in the lungs than the vector control cells (Supplementary Figure S5J-K). These results suggested that colonic PDAP1 may function in tumor initiation or as a neoplastic factor that contributes to CRC progression.
3.3 Pdap1 deficiency impairs mucosal restitution and retards tumor initiation and growth
As PDAP1 is highly conserved between humans and mice (Supplementary Figure S6A), we generated Villin-Cre;Pdap1fl/fl mice with intestinal epithelial-specific knockout of Pdap1, which appeared to develop normally (Supplementary Figure S6B). In the DSS-induced colitis model, Villin-Cre;Pdap1fl/fl mice appeared to be markedly more sensitive to the injurious effects of DSS. Weight loss and contraction in colon length were more pronounced in Villin-Cre;Pdap1fl/fl mice (Figure 1A,B). Accordingly, Villin-Cre;Pdap1fl/fl mice showed increased dextran permeability, strongly suggesting epithelial barrier dysfunction (Figure 1C). Flow cytometry analysis showed that DSS-treated Villin-Cre;Pdap1fl/fl mice exhibited increases in neutrophils, macrophages, and CD4+ T cell infiltration (Figure 1D) and different patterns of chemokine and cytokine expression when compared to the control mice (Figure 1E). Histological examination confirmed the presence of multiple erosions and intense inflammatory changes, including crypt abscesses. Although the control mice exposed to DSS showed evidence of mucosal erosions, most of these were small and exhibited features typical of mucosal healing, with complete re-epithelialization of most lesions. In contrast, Villin-Cre;Pdap1fl/fl mice exposed to DSS lacked evidence of re-epithelialization (Figure 1F). Consistent with these results, BrdU and TUNEL staining confirmed that the lack of PDAP1 resulted in impaired epithelial proliferation and increased apoptosis (Figure 1F). Furthermore, RNA sequencing (Figure 1G) and gene set enrichment analysis (GSEA) showed that gene set DNA replication, cell adhesion molecules, cytokine-cytokine receptor interaction, cell cycle, focal adhesion, ECM receptor interaction, chemokine signaling pathway, and mitogen-activated protein kinase (MAPK) signaling pathway were upregulated in the control mice (Figure 1H). These results suggest that intestinal knockout of Pdap1 impaired the defense of the intestinal mucosa in mice.
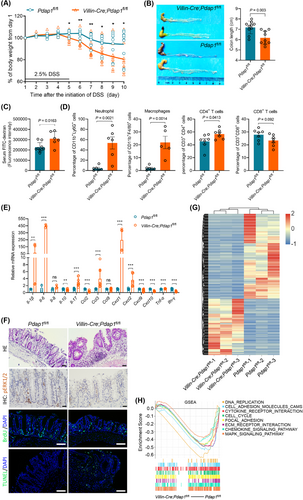
Furthermore, an AOM/DSS model was established to evaluate the role of Pdap1 in the initiation and development of CAC. Consistent with the results from human CRC tissues, PDAP1 expression was elevated in CAC tissues compared to that in the normal mucosa (Figure 2A). There was greater body weight loss in Villin-Cre;Pdap1fl/fl mice (Figure 2B), which was consistent with that observed in DSS-induced colitis. Moreover, we found that the number and size of tumors were significantly decreased in Villin-Cre;Pdap1fl/fl mice compared with control mice (Figure 2C-E). Flow cytometric analysis showed that tumors from Villin-Cre;Pdap1fl/fl mice exhibited decreased macrophage infiltration (Figure 2F). Histological examination confirmed the presence of colonic carcinoma and BrdU staining confirmed that the absence of PDAP1 resulted in impaired cell proliferation (Figure 2G). These results suggested that intestinal Pdap1 loss decreased CAC initiation and development.
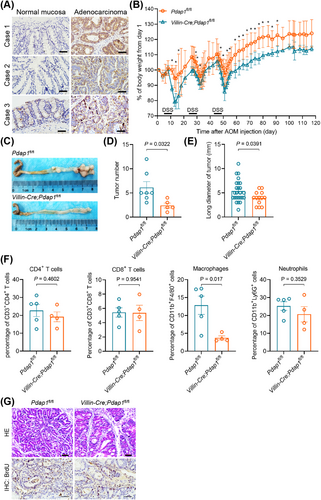
Loss of Pdap1 inhibits tumor initiation and growth in AOM/DSS colitis-associated carcinoma model. (A) Representative images of IHC staining of PDAP1 in AOM/DSS-induced colorectal carcinoma tissues and paired normal mucosa. Scale bar, 200 μm. (B) Weight loss (expressed as a percentage of the initial weight) of Villin-Cre;Pdap1fl/fl and Pdap1fl/fl mice following 2% DSS treatment over time. (C) Representative macroscopic views of colons from mice of the indicated genotypes. (D-E) Quantification of tumor number (D) and tumor long diameter (E). Tumors were harvested from Villin-Cre;Pdap1fl/fl and Pdap1fl/fl mice following AOM/DSS treatment on day 120. (F) Quantification of the infiltrated immunocytes in tumors detected by flow cytometry analysis. (G) Representative images of HE and IHC staining of BrdU in tumors. Scale bar, 50 μm. The Pvalues in (B) and (D-E) were determined using two-tailed Student's t-test. *, P < 0.05.
Abbreviations: AOM, azoxymethane; DSS, dextran sulfate sodium; HE, hematoxylin and eosin stain; IHC, immunohistochemistry; BrdU, 5-bromo-2′-deoxyuridine.
3.4 PDAP1 promotes FRA-1 expression
To explore the mechanisms by which PDAP1 promotes CRC progression, we used gene expression profiling in three PDAP1 knockdown CRC cell lines (Figure 3A). Concordant with its effects on DNA replication in the S phase, GSEA revealed a positive association between the PDAP1 profile and DNA replication signature (Figure 3B). Of the 913 DEGs identified, FOSL1, which encodes Fos-related antigen 1 (FRA-1), was a unique differentially expressed gene in all three PDAP1 knockdown cell lines compared to paired control cells (Figure 3C). The downregulation of FOSL1 by PDAP1 knockdown was confirmed by qPCR (Figure 3D) and western blotting (Figure 3E). The correlation between PDAP1 and FOSL1 was further validated using GEO datasets (Supplementary Figure S7A). We also found that overexpression of PDAP1 in CRC cells increased FOSL1 expression at both the mRNA (Supplementary Figure S7B) and protein (Supplementary Figure S7C) levels. Further analysis showed that PDAP1 mildly affected the turnover (Supplementary Figure S7D), proteasome-mediated degradation (Supplementary Figure S7E), and cellular localization (Figure 3F and Supplementary Figure S7F) of FRA-1. Furthermore, we analyzed PDAP1 and FRA-1 expression in serial sections of clinical CRC tissues (Figure 3G) and found that FRA-1 expression was significantly correlated with PDAP1 expression (ρ = 0.481, P = 0.00003, Supplementary Table S3). In addition, we confirmed the regulatory role of PDAP1 in FRA-1 expression in a panel of hepatocellular carcinoma cells and found that overexpression of PDAP1 led to increased FRA-1 expression in all the checked cells (Supplementary Figure S7G). Using TCGA data, we found a positive correlation between PDAP1 and FOSL1 in many cancers and normal tissues (Supplementary Table S4). These results indicated that PDAP1 promotes FRA-1 expression in CRC.

PDPA1 promotes FRA-1 expression. (A) Heatmap of DEGs in control (shCtrl) and PDAP1 knockdown (shPDAP1) CRC cells. (B) GSEA of gene expression changes in SW480 shPDAP1 cells compared with SW480 shCtrl cells. (C) Venn diagram depicting the overlap of DEGs from (A). (D) qPCR analysis of PDAP1 and FOSL1 in the indicated CRC cells transfected with shCtrl or shPDAP1. (E) Western blot analysis of FRA-1 and PDAP1 in the indicated CRC cells transfected with shCtrl or shPDAP1. The graph shows semi-quantitative analysis of relative FRA-1 expression. (F) Representative images of immunofluorescence staining of FRA-1 and PDAP1 in HCT-8 cells transfected with control plasmid or PDAP1 expression plasmid. Scale bar, 50 μm. (G) Representative images of immunohistochemical staining of PDAP1 and FRA-1 in human CRC tissues. The heatmap shows IHC score. The P-values in (D-E) were determined using two-tailed Student's t-test.
Abbreviations: KEGG, Kyoto Encyclopedia of Genes and Genomes; NES, normalized enrichment score; FDR, false discovery rate; shCtrl, scramble control; shPDAP1, knockdown of PDAP1; IHC, immunohistochemistry.
3.5 PDAP1 promotes FRA-1 expression regardless of its phosphorylation or sumoylation status
PDAP1 can be phosphorylated by casein kinase II [11], and numerous phosphoproteomic studies have identified several phosphorylation modification sites in multiple tissues [24]. We selected the eight most frequently phosphorylated sites and mutated these residues to alanine. As shown in Supplementary Figure S8A, these mutations did not significantly affect FRA-1 expression. Furthermore, PDAP1 was predicted to undergo sumoylation modification [25]. We mutated the potential sumoylation sites to alanine and found that these mutations had little effect on FRA-1 expression (Supplementary Figure S8B). These results indicate that PDAP1-induced FRA-1 expression was not dependent on its phosphorylation or sumoylation modifications.
3.6 PDAP1 promotes CRC progression via FRA-1
Because FRA-1 was regulated by PDAP1, we investigated whether FRA-1 mediated PDAP1-induced CRC progression. We found that overexpression of FRA-1 in PDAP1 knockdown cells (Figure 4A) reversed the effects of PDAP1 knockdown on colony formation (Figure 4B), cell invasion (Figure 4C), migration (Figure 4D), proliferation (Figure 4E), and cell cycle progression (Figure 4F). In vivo assays showed that the recovery expression of FRA-1 in PDAP1 knockdown cells restored the growth and metastatic abilities of PDAP1 knockdown cells (Figure 4G,H). These results indicated that PDAP1 facilitated migration, invasion, proliferation, cell cycle progression, and metastasis, mainly by stimulating FRA-1 expression.
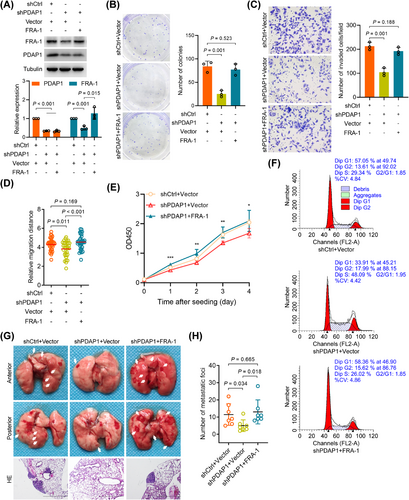
PDAP1 promotes proliferation, invasion, and metastasis via FRA-1. (A) Western blot analysis of FRA-1 and PDAP1 in HCT-8 cells transfected with control plasmid, shPDAP1 alone or in combination with FRA-1 expression plasmid. The graph shows semi-quantitative analysis of relative PDAP1 and FRA-1 expression. (B) Clonogenic assay of HCT-8 cells transfected with control plasmid, shPDAP1 alone or in combination with FRA-1 expression plasmid. The graph shows the colony number. (C) Representative images of HCT-8 cells invading through Matrigel-coated transwell inserts toward serum for 24 h. HCT-8 cells were transfected with control plasmid, shPDAP1 alone or in combination with FRA-1 expression plasmid. Scale bar, 50 μm. The graph shows the average number of invaded cells per field. (D) Quantification of the migration ability of HCT-8 cells transfected with control plasmid, shPDAP1 alone or in combination with FRA-1 expression plasmid. (E) Proliferation curve of HCT-8 cells transfected with control plasmid, shPDAP1 alone or in combination with FRA-1 expression plasmid determined using CCK-8 assays. (F) Flow cytometry analysis of cell cycle progression of HCT-8 cells transfected with control plasmid, shPDAP1 alone or in combination with FRA-1 expression plasmid. (G) Representative images of lung metastases and HE staining of metastatic tumors harvested from mice injected with the indicated cells on day 52. Scale bar, 2mm. (H) Quantification of lung metastases from each mouse injected with the indicated cells, and data are presented as a scatter diagram. The P-values in (A-E) and (H) were determined using two-tailed Student's t-test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Abbreviations: shCtrl, scramble control; shPDAP1, knockdown of PDAP1; HE, hematoxylin and eosin stain.
3.7 PDAP1 interacts with EGFR and promotes EGFR signaling
To investigate the underlying mechanism by which PDAP1 induces FRA-1 expression, we analyzed the intracellular signaling using a phosphoprotein antibody array (Supplementary Figure S9A). Of 304 proteins involved in the 16 cellular pathways analyzed, the most significant effects of PDAP1 overexpression were to induce phosphorylation of MAPK/ERK kinase 1 (MEK 1) (Figure 5A) and regulate MAPK signaling (Figure 5B). These results were confirmed by western blotting against ERK1/2 phosphorylated isoforms (T202/Y204) compared to total ERK1/2 protein (Supplementary Figure S9B). Furthermore, we characterized the interacting partners of PDAP1 using a His pull-down assay and LC-MS/MS (Supplementary Table S5). Of the 38 potential interacting proteins identified, epidermal growth factor receptor (EGFR), which is closely linked to the Raf/MAPK/ERK1/2 signaling pathway, was selected for further analysis. We confirmed the interaction between PDAP1 and EGFR, using co-immunoprecipitation (Supplementary Figure S9C). Immunofluorescence staining showed that PDAP1 co-localized with EGFR along the cytosolic side of the cell membrane in both HCT-8 and human CRC tissues (Figure 5C and Supplementary Figure S9D). To determine which domain of EGFR was responsible for interacting with PDAP1, we truncated EGFR and fused a GFP tag to the carboxyl terminus (Figure 5D). As shown in Figure 5E, the intracellular juxtamembrane (JM) region of EGFR is indispensable for its interaction with PDAP1. Truncation analysis of PDAP1 based on its predicted structure (Supplementary Figure S9E) showed that the C-terminal domain of PDAP1 was responsible for its interaction with EGFR (Supplementary Figure S9F-G). We further analyzed the effect of PDAP1 on EGFR activation. We found that PDAP1 knockdown impaired the phosphorylation of EGFR at Tyr1173 and Tyr1086 and ERK1/2 at Thr202 and Tyr204 in EGF-treated CRC cells (Figure 5F and Supplementary Figure S9H). In contrary, overexpression of PDAP1 enhanced the phosphorylation of EGFR at Tyr1173 and Tyr1086 and ERK1/2 at Thr202 and Tyr204 in EGF-treated CRC cells (Figure 5G). These results suggested that PDAP1 interacts with the JM domain of EGFR and promoted EGFR/MAPK signaling.
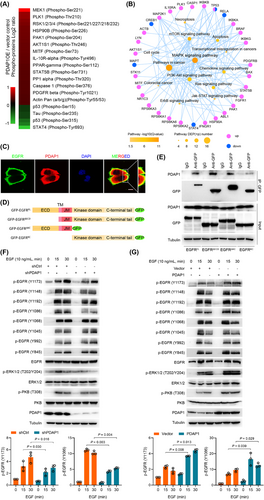
PDAP1 interacts with EGFR and promotes EGFR signaling. (A) Heatmap shows differentially expressed phosphosites in HCT-8 cells overexpressing PDAP1 compared with control cells. (B) Signaling network analysis of differentially expressed phosphosites in HCT-8 cells overexpressing PDAP1 compared with control cells. (C) Representative images of immunofluorescence staining of endogenous PDAP1 and EGFR in HCT116. Scale bar, 50 μm. (D) Schematic representation of the EGFR constructs used in this study. (E) Western blot analyses of endogenous PDAP1 co-IP with EGFR. HCT-8 cells were transfected with the indicated constructs as shown in (D). (F) Western blot analyses of the indicated proteins in HCT-8 shCtrl or HCT-8 shPDAP1 cells after increasing periods of EGF treatment. The graph shows semi-quantitative analysis of relative p-EGFR (Y1173) and p-EGFR (Y1086) expression. (G) Western blot analyses of the indicated proteins in HCT-8 cells transfected with control plasmid or PDAP1 after increasing periods of EGF treatment. The graph shows semi-quantitative analysis of relative p-EGFR (Y1173) and p-EGFR (Y1086) expression. The P-values in (F-G) were determined using two-tailed Student's t-test.
Abbreviations: co-IP, co-immunoprecipitation; DAPI, 4′,6-diamidino-2-phenylindole; ECD, extracellular domain; TM, transmembrane domain; JM, juxtamembrane.
3.8 PDAP1 promoted FRA-1 via EGFR/MAPK signaling
As previous results indicated transcriptional regulation of FRA-1 by PDAP1 (Figure 3D and Supplementary Figure S7B), we confirmed this regulation in HCT-8 cells by a luciferase reporter assay (Supplementary Figure S10A). Given that PDAP1 enhances EGFR-MAPK signaling (Figure 5) and Ras-MAPK signaling regulates FRA-1 in multiple contexts [26-28], we examined the involvement of EGFR-MAPK signaling in PDAP1-induced FRA-1 expression, by treating PDAP1-overexpressing cells with the phosphatidylinositol-3 kinase (PI3K) inhibitor ZSTK474 or MEK1 inhibitor GDC-0623. We found that PDAP1 overexpression enhanced the activation of protein kinase B (PKB) and ERK1/2; MEK1 inhibition, but not PI3K inhibition, eliminated the upregulation of FRA-1 induced by PDAP1 overexpression in both HCT-8 (Supplementary Figure S10B) and HCT116 (Supplementary Figure S10C) cells. Further analysis using the EGFR inhibitor PD153035 showed that EGFR inhibition eliminated the effects of PDAP1 overexpression on the activation of PKB and ERK1/2 and the upregulation of FRA-1 in a dose-dependent manner (Supplementary Figure S10D). These results suggested that PDAP1 induced FRA-1 expression via EGFR/MAPK signaling.
3.9 c-Myc directly transactivates PDAP1 in CRC
To investigate the mechanism underlying the upregulation of PDAP1 in CRC cells, we first analyzed copy number alterations (CNA) in CRC using TCGA data. Analysis of the PDAP1 CNA distribution showed that amplification represented only a small proportion of the samples (Supplementary Figure S11A). Furthermore, when we quantified the PDAP1 gene copy number in DNA from 10 primary CRC tissues and paired adjacent mucosa we observed no significant difference (Supplementary Figure S11B-C). These results indicated that PDAP1 overexpression is not driven by PDAP1 CNA. Next, we constructed a series of 5’-truncated versions of the PDAP1 promoter and found that the promoter region between -254 bp and -1 bp was indispensable for basal transcription of PDAP1 in CRC cells (Supplementary Figure S12A and Figure 6A). Sequence analysis identified two c-Myc binding sites in the promoter between -254 bp and -1 bp (Figure 6B). ChIP-qPCR confirmed the direct binding of c-Myc and PDAP1 promoter (Figure 6C). Further analysis showed that mutation of the c-Myc binding sites, especially the first binding site, decreased reporter activity (Figure 6D), indicating that PDAP1 transcription is primarily regulated by c-Myc. These results were further confirmed by the overexpression of stable c-Myc (c-MycT58A) and shRNA-mediated knockdown of c-Myc in CRC cells, which resulted in increased and decreased PDAP1 expression, respectively (Figure 6E,F and Supplementary Figure S12B-C). In addition, inhibiting c-Myc with 10058-F4 blocked PDAP1 expression in a dose-dependent manner (Figure 6G-H and Supplementary Figure S12D). Gene expression analysis using TCGA data showed that PDAP1 expression was highly correlated with MYC expression in CRC (Supplementary Figure S12E). Consistently, the analysis of c-Myc and PDAP1 expression in 282 CRC tissues using immunohistochemistry (IHC) staining showed that they were significantly correlated (Figure 6I). These results indicated that c-Myc-mediated transactivation of PDAP1 contributes to its overexpression in CRC.
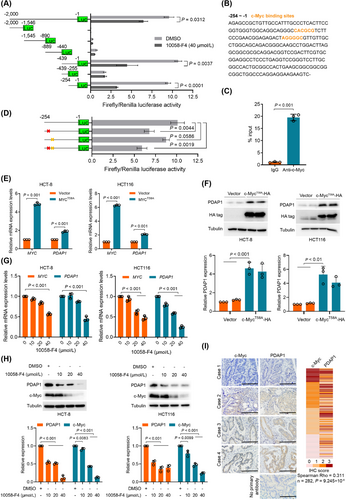
c-Myc directly transactivates PDAP1 in CRC. (A) Schematic representation of the PDAP1 promoter reporter constructs. HCT-8 cells were transfected with the indicated constructs and treated with DMSO or 10058-F4. (B) Sequence of the PDAP1 promoter spanning -254 to -1 base pairs (bp). Orange letters indicate putative c-Myc binding sites. (C) ChIP assay for the enrichment of c-Myc at the PDAP1 promoter in HCT-8 cells. ChIP-qPCR values are expressed as median of percentage chromatin immunoprecipitated, normalized to the isotype-matched control. The P-value was determined using an unpaired Mann-Whitney test. (D) Schematic representation of the PDAP1 promoter reporter constructs. HCT-8 cells were transfected with the indicated constructs. Red cross, mutation of the first c-Myc binding site; yellow cross, mutation of the second c-Myc binding site. (E) qPCR for MYC and PDAP1 mRNA expression normalized to GAPDH mRNA expression in HCT-8 or HCT116 cells. The cells were transfected with control plasmid or MYCT58A expression plasmid. (F) Western blot analyses of the indicated proteins in HCT-8 or HCT116 cells transfected with control plasmid or MYCT58A expression plasmid. The graph shows semi-quantitative analysis of relative PDAP1 expression. (G) qPCR for MYC and PDAP1 mRNA expression normalized to GAPDH mRNA expression in HCT-8 or HCT116 cells. The cells were treated with increasing amounts of 10058-F4. (H) Western blot analyses of the indicated proteins in HCT-8 or HCT116 cells. The cells were treated with increasing amounts of 10058-F4. The graph shows semi-quantitative analysis of relative PDAP1 and c-Myc expression. (I) Representative images of immunohistochemical staining of c-Myc and PDAP1 in human CRC tissues. The heat map shows the IHC score. The P-values in (A) and (C-H) were determined using two-tailed Student's t-test.
Abbreviations: bp, base pairs; ChIP, chromatin immunoprecipitation; Luc, luciferase; IHC, immunohistochemistry.
3.10 Depletion of PDAP1 reduced tumor growth in vivo
Having shown that PDAP1 promotes CRC progression, we then investigated the potential of targeting PDAP1 to prevent tumor growth in vivo. In a PDX model (Figure 7A), depletion of PDAP1 using in-vivo-optimized siRNAs targeting PDAP1 significantly reduced FRA-1 expression and tumor growth (Figure 7B-E). These results demonstrated that inhibition of PDAP1 inhibited FRA-1 expression and cancer development, highlighting its role as a potential therapeutic target.
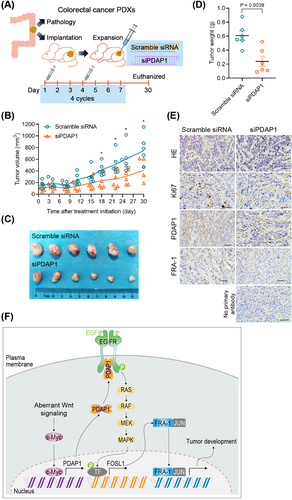
Treatment of CRC PDXs with siRNAs targeting PDAP1 reduces tumor growth. (A) Schematic representation of CRC PDX model, as well as the schedule of scramble siRNA and siRNAs targeting PDAP1 (siPDAP1) administration. After PDX implantation, the mice were randomized to two treatment groups (n = 6 in each group). (B) Tumor volume of CRC PDXs treated with scramble siRNA or siPDAP1. (C) Images of tumors formed after scramble siRNA or siPDAP1 treatment of CRC PDXs. (D) The graph shows quantification of tumor weight of CRC PDXs treated with scramble siRNA or siPDAP1. (E) Representative images of HE and immunohistochemical staining of Ki67, PDAP1, and FRA-1 in CRC PDXs after scramble siRNA or siPDAP1 treatment. (F) Model depicting the proposed mechanism mediating CRC development. The Pvalues in (B) and (D) were determined using two-tailed Student's t-test. *, P < 0.05.
Abbreviations: PDX, patient-derived xenograft; siPDAP1, siRNAs targeting PDAP1.
4 DISCUSSION
Although we and others have shown that PDAP1 is overexpressed in several types of cancers, including rectal cancer [14-16], the origin of PDAP1 overexpression in cancer is poorly understood. Using CNA analysis, we demonstrated that PDAP1 overexpression is unlikely to result from PDAP1 gene amplification, which is distinct from the previous observation that PDAP1 is amplified in a hepatocellular carcinoma cell line [29]. The oncoprotein MYC is a master regulator of gene transcription and a potent driver of transformation. In non-transformed intestinal epithelial cells, MYC-regulated transcription is highly regulated; however, in transformed intestinal epithelial cells, MYC is often dysregulated through constitutive and elevated MYC expression, which occurs through a wide variety of mechanisms, including MYC gene amplification [30] and aberrant Wnt signaling [7, 8], and enables MYC to initiate and/or sustain tumor growth. Here, we revealed that PDAP1 is a novel target of c-Myc and facilitates CRC initiation and progression by activating EGFR-MAPK-FRA-1 signaling (Figure 7F), indicating that PDAP1 mediates the crosstalk between MYC signaling and EGFR-MAPK signaling.
EGFR signaling is a known mediator of colorectal carcinogenesis, and thus it is an important target for several cancer therapeutics currently in clinical use. However, only a subgroup of patients benefit from these treatments [31]. The detailed picture of dysregulated EGFR signaling remains unclear and needs to be elucidated to understand CRC pathogenesis and to develop efficient treatments. The 38 amino acid cytoplasmic JM region of EGFR includes receptor-trafficking signals [32-34] and is required for kinase activation [35-37]. Structural analysis reveals that the C-terminal half of the JM segment latches the activated kinase domain to the activator, and the N-terminal half of this segment further potentiates dimerization, most likely by forming an antiparallel helical dimer that engages the transmembrane helices of the activated receptor [38]. Using deletion mutations, we found that PDAP1 interacts with the JM region of EGFR and promotes EGF-induced EGFR phosphorylation at Y1173 and Y1086. One possible explanation is that PDAP1 binding stabilizes the active conformation. However, conclusive evidence of EGFR conformational changes induced by PDAP1 binding depends on structural studies.
Inflammatory bowel disease involves an uncontrolled immune-mediated inflammatory response and damage to the gastrointestinal epithelium in genetically predisposed individuals to an unknown environmental trigger that interacts with the intestinal flora [39]. Given that EGFR signaling plays a central role in the regulation of colon epithelial biology and the response to injury and inflammation, it is not surprising that conditional knockout of Pdap1, a coactivator of EGFR, in mouse intestinal epithelial cells led to impaired restitution and worsened outcomes of colitis, which is consistent with observations in mice with Egfr deficiency [40, 41].
In the AOM/DSS-induced CAC model, loss of PDAP1 in intestinal epithelial cells results in increased weight loss after each round of DSS treatment but inhibits carcinogenesis, which is consistent with the observation that diminished EGFR signaling in Egfrwa2/wa2 mice or treatment with pharmacological EGFR inhibitors reduces tumor formation [42, 43]. However, different studies have also supported a protective role for EGFR in CRC. It has been shown that reduced EGFR signaling in the antimorphic Egfrwa5/+ or hypomorphic Egfrwa2/wa2 background augments colitis severity and accelerates and increases tumor development in the Il10−/− mouse model of CAC [44, 45]. Furthermore, AOM/DSS-induced CAC is more invasive in Egfrwa5/+ mice [41]. This discrepancy may be because the influence of EGFR on tumorigenesis depends on the cell type from which it is expressed and the experimental conditions [46, 47].
Activator protein-1 (AP-1) is a transcription factor that consists of either a Jun-Jun homodimer or a Jun-Fos heterodimer. AP-1 regulates the expression of multiple genes essential for cell proliferation, differentiation and apoptosis [48]. FRA-1, encoded by FOSL1, belongs to the FOS protein family and mainly forms an AP-1 complex with proteins of the JUN family to regulate human tumor progression and metastasis [49], thus representing a promising therapeutic target. FRA-1 is overexpressed in many tumors, including CRC [50] and other cancers [49], and controls cell mobility, invasion, and proliferation in various cancers, including breast cancer [51] and CRC [52]. The present study revealed that PDAP1 is a novel regulator of FRA-1 expression and that the close correlation between PDAP1 and FRA-1 is conserved in various cancers and normal tissues, indicating that PDAP1 is an intrinsic regulator of FRA-1. As expected, in vitro and in vivo assays have demonstrated that FRA-1 mediates the oncogenic function of PDAP1. It is believed that the regulation of FRA-1 expression is multifaceted, and the MAPK pathway is the most prominent among the upstream regulatory pathways of FRA-1 [53]. In accordance with this observation, we demonstrated that MAPK signaling mediates PDAP1-induced FRA-1 expression in CRC cells. Through signaling pathway screening, we also showed that PDAP1 regulates other signaling pathways besides MAPK signaling. Further studies are needed to elucidate the effects and mechanisms by which PDAP1 regulates these signaling pathways.
We acknowledge several limitations in the present study. First, since CAC cell model is lacking, the effects of PDAP1 on tumor cells were only measured in sporadic colon cancer cells. Second, we did not investigate the role of PDAP1 in CRC initiation and progression using ApcMin/+ mice. Third, we only investigated short-term effects of PDAP1 knockout on intestinal inflammation in the animal model. Further studies are needed to fully elucidate other functions of PDAP1 in CRC. Our pathway enrichment analysis revealed that PDAP1 affects several important oncogenic pathways. The effects of these pathways, including the p53 signaling pathway, PPAR signaling pathway, propanoate metabolism, steroid biosynthesis, and spliceosome, are still largely uncharacterized. This is possibly due to the multilayered and complex role of PDAP1 in tumor cells.
5 CONCLUSIONS
This study outlines the pro-oncogenic role of c-Myc-driven PDAP1 overexpression in CRC initiation and progression, which is mediated by the EGFR-MAPK-FRA-1 signaling axis. Moreover, our in vitro and in vivo results provided a rationale for the development of new therapeutic agents targeting PDAP1 for CRC treatment.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study was approved by the Hospital Ethics Committee of Xijing Hospital (permit number: KY20213194-1). Written informed consent was obtained from all participants. All animal protocols were approved by the Animal Care and Welfare Committee of the Fourth Military Medical University (permit number: 20220861).
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data that support the findings of this study are available from the corresponding author upon reasonable request.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
Hong-Yong Cui and Shi-Jie Wang conceived and designed the project. Wei Wei, Mei-Rui Qian, Ruo-Fei Tian and Xin Fu performed experiments and/or conducted data acquisition and analyses; Hong-Wei Li, Gang Nan, Ting Yang, Peng Lin, Jian-Hua Dou, Xi Chen, Yu-Meng Zhu, Bin Wang and Xiu-Xuan Sun contributed technical/reagents materials, analytic tools, and/or grant support; Hong-Yong Cui, Shi-Jie Wang, Jian-Li Jiang, Ling Li and Zhi-Nan Chen prepared, wrote, reviewed, and/or revised the manuscript.
ACKNOWLEDGEMENTS
Not applicable.




