ZBED2 expression enhances interferon signaling and predicts better survival of estrogen receptor-negative breast cancer patients
Dingxie Liu and Qiongyu Hao contributed equally to the study.
Abbreviations
-
- BC
-
- breast cancer
-
- CSCs
-
- cancer stem cells
-
- DMFS
-
- distant-metastasis-free survival
-
- ER
-
- estrogen receptor
-
- FPKM
-
- Fragments Per Kilobase of transcript sequence per Millions base pairs sequenced
-
- GO
-
- Gene Ontology
-
- GSEA
-
- Gene Set Enrichment Analysis
-
- HER2
-
- human epidermal growth factor receptor-2
-
- HR
-
- hazard ratio
-
- IFN
-
- interferon
-
- IHC
-
- immunohistochemistry
-
- IRF9
-
- IFN regulatory factor 9
-
- ISGs
-
- interferon stimulated genes
-
- OS
-
- overall survival
-
- PDA
-
- pancreatic ductal adenocarcinoma
-
- PFS
-
- progression-free survival
-
- PR
-
- progesterone receptor
-
- RFS
-
- relapse-free survival
-
- shRNAs
-
- short-hairpin RNA
-
- STAT
-
- signal transducer and activator of transcription
-
- TFs
-
- transcription factors
-
- TNBC
-
- triple-negative BC
The molecular pathogenesis of estrogen receptor (ER)-negative, especially triple-negative breast cancer (TNBC), is unclear, thus, the treatment of these types of cancers is still a great challenge [1]. Chemotherapy remains the primary adjuvant treatment choice for TNBC patients [2], but a large number of these tumors are resistant to chemotherapy and relapse or metastasize quickly after adjuvant therapy [3]. Hence, TNBC is still a disease related to poor prognosis and limited treatment options. However, tumor lymphocyte infiltration in the TNBC microenvironment is related to good prognosis and chemotherapy response [4]. To avoid over-treatment, it is imperative to identify reliable markers to predict outcome and treatment response in TNBC patients.
The activation of immune response genes has been proved to be a good prognostic factor for ER-negative cancers [5]. Previously, we [6, 7] and others [8, 9] have identified some prognostic gene signatures for ER-negative, triple-negative, and basal-like BCs. However, more sensitive and effective prognostic biomarkers for these cancer subtypes are in urgent need.
Transcription factors (TFs) are key regulators of many biological processes, including cell differentiation and proliferation. TFs are situated in the hubs of a complex network shaping the hallmarks of human cancer, and aberrant epigenetic/genetic alterations of TFs are involved in the tumorigenesis of many cancers [10]. The expression of many TFs was found to be associated with patient survival in various cancers [10]. A deeper understanding of TF regulation opens up new possibilities for using TFs as targets of anticancer drugs or biomarkers for predicting prognosis. Here, we examined the association of 1198 TFs with the prognosis of patients with ER-negative BC. Eight independent BC cohorts (n = 13,434) were analyzed. Gene functions were predicted using Bayesian binary regression and gene set enrichment approaches, and were further validated by in vitro functional studies. Our aim was to identify TFs associated with the molecular pathogenesis of ER-negative BC and to provide a more specific classification of clinical significance.
Cox regression analysis of these 1198 genes in four U133-based cohorts (cohorts 2, 3, 4 and CIT) showed that 42 genes were significantly associated with the recurrence of ER-negative BCs (Supplementary Figure S1A, Supplementary Table S1). Among the 42 TF genes, 25 genes including interferon regulatory factor (IRF)1, 2, 8 & 9 and signal transducer and activator of transcription (STAT)1 & 4 were annotated under the inflammation/immune-related functional terms (Supplementary Figure S1B). Interestingly, these 25 genes were all favorable factors for the relapse-free survival of ER-negative BC patients (Supplementary Figure S1A). These data suggested that TF genes that regulate inflammation/immune signaling may have important roles in ER-negative BC.
We were particularly interested in the Zinc Finger BED-Type Containing 2 (ZBED2) gene because it is the most significantly associated gene with the survival of ER-negative BC patients among the 42 candidate TF genes (Supplementary Figure S1A). To validate whether these ZBED2 Affymetrix array-based results could be repeated in cohorts from other array platforms, three additional BC cohorts were included and a total of 13,412 cases (11,146 cases have survival information) were tested in this study (Supplementary Table S2). Univariate Cox analysis showed that ZBED2 expression was consistently associated with better survival of ER-negative BC but not in ER-positive BC in all the 7 cohorts (Supplementary Figure S2A-B). ZBED2 expression was also consistently associated with better outcome in TNBC and basal-like BC in most of the 7 cohorts (Supplementary Figure S2C-D). Although ZBED2 expression was also a strong predictor for human epidermal growth factor receptor2 (HER2)-enriched BC, interestingly, it was only weakly associated with survival of HER2-positve BC patients in most of the 7 cohorts (Supplementary Figure S2E-F).
Multivariate regression analysis showed that after adjusting for different clinical variables in all seven cohorts, the association of ZBED2 expression with survival remained significant in all ER-negative and some basal-like BC subcohorts (Supplementary Figure S3, Supplementary Table S3), suggesting that ZBED2 expression is an independent prognostic factor of BC. The significant impact of ZBED2 expression on ER-negative BC and TNBC outcomes was also supported by Kaplan-Meier analysis (Supplementary Figure S4).
To unravel the potential biological processes associated with ZBED2, we analyzed 27 tumor-related pathways and found that ZBED2 expression was most associated with immune-related signal pathways, including interferon α (IFNα), IFNγ and tumor necrosis factor α (TNFα) pathways in BC, particularly in ER-negative, triple-negative, basal-like and HER2-enriched BCs (Figure 1A, Supplementary Figures S5-S6). Interestingly, the expression of ZBED2 in these BC subtypes was similarly higher than that of ER-positive and Luminal BCs (Supplementary Figure S7).
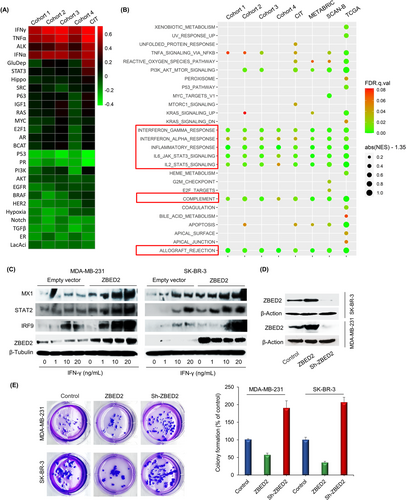
Gene set enrichment analysis (GSEA) showed that 12 hallmark gene sets were significantly enriched in ER-negative BCs with high ZBED2 expression in at least 3 cohorts, and 7 gene sets were enriched in all the 7 cohorts (Figure 1B, Supplementary Table S4). These 7 gene sets are all related to immune/inflammation response. Notably, compared to the other enriched gene sets, the two IFN gene sets had a much higher percentage of genes contributing to the enrichment score (i.e., higher leading edge subset) (Supplementary Figures S8-S9, Supplementary Table S4). Overall, these data suggested that ZBED2 may be involved in immune/inflammation processes, most likely in IFN signaling in ER-negative BCs.
To validate the effect of ZBED2 on the IFN pathway, the effects of ZBED2 on the expression of several key components of IFN pathways in IFNγ-treated BC cells were analyzed using Immunoblotting. We demonstrated that specific interferon-stimulated genes (ISGs), STAT2, MX dynamin-like GTPase 1 (MX1), and IRF9, were induced as expected in response to IFN treatment, and the expression induction was significantly exacerbated by forced expression of ZBED2 in ER-negative BC cell lines MDA-MB-231 and SK-BR-3 (Figure 1C). These results indicated that ZBED2 functions as a stimulator of a specific subset of ISGs.
To further substantiate the cancer-relevant function of ZBED2, we performed clonogenic assays following ZBED2 overexpression or knockdown in both cell lines. ZBED2 knockdown using short-hairpin RNAs (shRNAs) resulted in a significant increase in anchorage-independent growth, whereas ZBED2 overexpression repressed growth significantly in both cell lines (Figures 1D and 1E). These results indicated that ZBED2 functions as a tumor suppressor through enhancing the IFN signaling in BC, at least in ER-negative BC or TNBC.
RNA-sequencing data from our ER-negative/triple-negative BC patient samples further confirmed that ZBED2 expression was positively correlated with expression of the above-mentioned ISGs (Supplementary Figure S10A-C). Samples with higher ZBED2 expression also had significantly higher IFN signaling activity (represented by IFN 6-gene score) than those with lower ZBED2 expression (Supplementary Figure S10D). Kaplan-Meier analysis of this in-house dataset showed that, similarly to IFN 6-gene signature, the expression of ZBED2 is a significant favorable survival factor in ER-negative BC (Supplementary Figure S10E-F).
Together, our research found that ZBED2 was related to immune response, supporting that TF genes that regulate inflammation/immune signaling play important roles in the molecular pathogenesis of ER-negative BC/TNBC. Additionally, we provide evidence that the effect of ZBED2 was related to promoting IFN signaling in ER-negative BC/TNBC. We also found that ZBED2 promoted the expression of IRF9 and improved the survival rate of TNBC patients. Therefore, the expression of ZBED2 and/or IRF9 can provide early insight into the prognosis of TNBC patients to identify those patients who are unlikely to respond to chemotherapy alone and may benefit from further immune-based treatment intervention.
In conclusion, ZBED2 can be used to predict the immune status of tumor and the risk of metastasis/recurrence before or during neoadjuvant chemotherapy to implement replacement therapy early and decrease the mortality of ER-negative BC/TNBC patients.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Compliance with ethical standard
CONSENT FOR PUBLICATION
Not applicable for this study.
ACKNOWLEDGMENTS
The results here are based upon the breast cancer microarray data from public repositories. We are very grateful to the investigators who contributed these microarray datasets to the public domain.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest, except that Liu has equity interest in Bluewater Biotech LLC.
AUTHOR CONTRIBUTIONS
DL and YW designed the study. DL, QH, JL, QL, KW, YW, JV, and YW performed the experiments and data interpretation. DL and YW wrote and edited the manuscript. All authors read and approved the final manuscript.
AVAILABILITY OF DATA AND MATERIALS
The datasets or codes used and/or analyzed during the current study are available from the corresponding author on reasonable request.
FUNDING
This work was supported in part by NIH-NIMHD U54MD007598, U54CA143931, NIH/NCI1U54CA14393, U56 CA101599-01; Department-of-Defense Breast Cancer Research Program grant BC043180, NIH/NCATS CTSI UL1TR000124 to J.V. Vadgama; Accelerating Excellence in Translational Science (AXIS) Pilot Grants G0812D05, NIH/NCI SC1CA200517 and 9 SC1 GM135050-05 to Y. Wu; NIH/NIMHD Accelerating Excellence in Translational Science (AXIS) Pilot Grants G0814C01 (Q. Hao).




