The repertoire of germline variants in patients with early-onset rectal cancer
Caroline Moraes Beltrami and Luisa Matos do Canto contributed equally to this work.
Abbreviations
-
- APC
-
- APC regulator of WNT signaling pathway
-
- ATM
-
- ATM serine/threonine kinase
-
- BMPR1A
-
- bone morphogenetic protein receptor type 1A
-
- BRCA2
-
- BRCA2 DNA repair associated
-
- CDKN2A
-
- cyclin-dependent kinase inhibitor 2A
-
- CHEK2
-
- checkpoint kinase 2
-
- CRC
-
- colorectal cancer
-
- EGFR
-
- epidermal growth factor receptor
-
- EORC
-
- early-onset rectal cancer
-
- FAP
-
- familial adenomatous polyposis
-
- HR
-
- homologous recombination
-
- KRAS
-
- Kirsten rat sarcoma viral oncogene homolog
-
- LP
-
- likely pathogenic
-
- LS
-
- Lynch syndrome
-
- MMR
-
- mismatch repair
-
- MSH2
-
- mutS homolog 2
-
- MUTYH
-
- mutY DNA glycosylase
-
- NF1
-
- neurofibromin 1
-
- P
-
- pathogenic
-
- PALB2
-
- partner and localizer of BRCA2
-
- PD-1
-
- programmed cell death 1
-
- POLD1
-
- DNA polymerase delta 1, catalytic subunit
-
- RB1
-
- RB transcriptional corepressor 1
-
- VUS
-
- variant of uncertain significance
The incidence of rectal cancer has increased in patients younger than 50 years old during the last decade. It is well established that young age at cancer onset is a hallmark of hereditary cancer. The prevalence of germline variants among early-onset rectal cancer (EORC) patients is largely unexplored. Here, we aimed to determine the spectrum of germline variants and their clinical impact in EORC patients diagnosed at age 40 or younger. We investigated 71 EORC patients (Supplementary Table S1), one of the largest cohorts to date, using a customized panel with 93 genes (Supplementary Table S2). A detailed description of the methods is provided as Supplementary Material.
We identified 94 germline variants in 52 of our patients (Figure 1A, Supplementary Table S3). Twenty-three patients presented 20 pathogenic (P) and/or eight likely pathogenic (LP) variants. High- and moderate-penetrance genes comprised most of these P/LP variants, mainly MSH2 (n = 5), MUTYH (n = 5), ATM (n = 5), and APC (n = 2) (Supplementary Figure S1). A family history of cancer was found in 56 of 71 patients, and 21 had a clinical diagnosis of Lynch Syndrome (LS). A hereditary colorectal cancer (CRC) predisposition syndrome was confirmed in 11 patients, of whom seven were carriers of P variants in MMR (mismatch repair) genes, two presented with P variants in APC, and two had MUTYH biallelic P/LP variants. However, this number increased to 13 since two variants of uncertain significance (VUS), one in APC and one in BMPR1A, were classified as P and LP (VarSome genetic database), respectively. A previous study detected seven P variants in known CRC predisposition genes (MMR genes and APC) in a cohort of 65 EORC patients, thus confirming the diagnosis in a similar proportion of patients than ours (∼11%) [1]. The MUTYH c.1187G>A monoallelic variant found in one patient was previously described [2], corroborating its impact on cancer emergence. Three patients with MMR-deficient tumors evaluated by immunohistochemistry also presented germline variants in MMR genes (Supplementary Table S3).
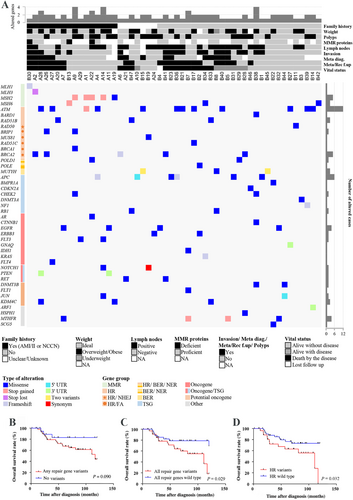
Among the patients with a family history of cancer and no clinical diagnosis of LS, variants in homologous recombination (HR) genes were the most prevalent (12 patients, including 5 with P/LP variants). Notably, 26 of our 52 patients with variants had alterations in DNA repair pathway genes. Although the involvement of variants in MMR genes with CRC risk is well defined, the effect of other repair genes remains unclear.
The most common altered genes were ATM (17%), APC (8%), MSH2 (7%), and BRCA2 (7%). Two ATM variants described here were previously reported in EORC patients (c.1229T>C and c.2932T>C) [3], and three patients showed the same ATM variant (c.8734A>G) previously described in CRC [4]. BRCA2 germline variants were also reported in EOCR patients [3]. A pan-cancer study identified ATM and BRCA2 germline variants in rectal cancer patients [2]. P and LP ATM variants were common in another pan-cancer study with 1,507 pediatric and young patients [5]. A previous study in CRC patients reported an enrichment of P variants in HR genes, particularly in ATM and PALB2 [6]. Germline variants in HR genes were described in young CRC patients [2, 7]. Other advanced tumors, such as pancreatic cancer, mesothelioma, and ovarian cancer, showed a tendency of enrichment of germline P variants in HR repair genes [8]. Taken together, these studies pointed out the relevance of germline variants of HR genes, particularly ATM and BRCA2, as risk factors for developing EORC.
Recently, a pan-cancer analysis reported P/LP variants of RB1 and NF1 in children, adolescents, and young adults with solid tumors [5]. Of note, we identified two patients with RB1 and one with NF1 variants. None of our cases developed retinoblastoma or had relatives with this cancer type. However, the presence of these variants highlights that novel discoveries depend on the selection criteria of genes investigated.
We conducted a co-segregation analysis in 17 relatives of seven patients (Supplementary Tables S4 and S5). Family members with cancer or polyps of three patients with P variants in known CRC predisposition genes (A1: MSH2; A4: APC; B15: biallelic MUTYH) had identical variants. Patients A1 and A4 also presented ATM variants shared by A4 relatives (classification of variants: VUS) but not by A1 relatives (classification of variants: P). For patient B45, both P/LP variants in MUTYH were found in two of four still unaffected relatives. Both patient B46 and one sister with thyroid cancer presented with CDKN2A VUS. Although this high-penetrance gene is associated mainly with hereditary melanoma, the same germline variant (c.170C>A) was previously identified in one CRC patient [9], thus supporting its contribution to the disease. Of interest, patient B29 presented a benign POLD1 variant (according to the ACMG criteria), and her sister with polyps also presented with the same variant. This variant could be associated with CRC appearance. Although co-segregation analysis provides additional evidence of an association between germline variants with cancer risk, functional assays are required to gain further insight into the potential pathogenic effect of candidate genetic variants.
In addition to predisposing individuals to cancer development, germline variants may impact outcomes. We found a lower overall survival in EORC patients presenting DNA repair gene variants than those without these variants (Figure 1B-D). Remarkably, a similar result was obtained considering only the patients with variants in HR genes.
Comparing our dataset with EORC data from six studies using next-generation sequencing, we found 16 commonly altered genes, of which ATM, APC, and MSH2 showed the highest number of germline variants (Supplementary Table S6 and Supplementary Figure S2).
The detection of germline variants is important not only for diagnosis and genetic counseling but also for determining therapeutic strategies. Particularly, patients with MMR variants could benefit from immune checkpoint inhibition (anti-PD-1 antibodies, such as pembrolizumab). Variants in HR genes (mainly BRCA1/2, CHEK2, and ATM) may indicate sensitivity to poly(ADP-ribose) polymerase inhibitors and platinum-based chemotherapy [10]. We found that two patients presented with CHEK2 variants. In a recent pan-cancer analysis with 11,947 patients and more than 50 types of cancer, P/LP variants in genes with therapeutic implications were found in 9% of patients [10]. Most of the germline variants with therapeutic actionability were identified in HR (BRCA1/2: 42%; CHEK2: 13%; and ATM: 12%) and MMR genes (11%) [10]. We also found germline variants in other genes with therapeutic relevance: EGFR, KRAS, and NF1. Advanced non-small cell lung cancers with EGFR mutations are treated with tyrosine kinase inhibitors. Currently, several KRAS inhibitors have been tested in solid tumors, including CRC. Sunitinib and selumetinib target therapies have been used in patients with NF1 germline variants.
Environmental factors (diet, obesity, and alterations in the gut microbiome) and low penetrance genetic variants may contribute to the pathogenesis of rectal cancer. In addition, the identification of germline variants in EORC patients has implications not only for diagnosis and genetic counseling but also for prognosis prediction and treatment selection. We identified a high frequency of EORC patients harboring germline variants (∼73%), mainly in HR/MMR genes, oncogenes, and tumor suppressors; of which some could be actionable targets for treatment. Screening programs should be implemented for all patients with EORC, regardless of their family history, and could be helpful to reverse the morbidity associated with the disease.
DECLARATIONS
ACKNOWLEDGMENTS
The authors would like to thank the patients and their family members for participating in this study. We are grateful to Dr. Rachel Pearlman and Dr. Heather Hampel from the Ohio University Comprehensive Cancer Center (Ohio, USA) for sharing their results in rectal cancer patients. Our thanks to the A.C.Camargo Cancer Center Biobank, São Paulo, Brazil. We acknowledge the technical assistance of Alexandre Defelicibus (A.C.Camargo Cancer Center) and Sandra Aparecida Drigo (Department of Surgery and Orthopedics, Experimental Research Unity, Faculty of Medicine, São Paulo State University - UNESP, Botucatu, SP, Brazil) during the development of this study.
FUNDING
This study was supported by grants from the Region of Southern Denmark Research Fund, Denmark, and the National Institute of Science and Technology in Oncogenomics (INCITO, FAPESP #2008/57887-9 and CNPq #573589/08-9), Brazil. Caroline Moraes Beltrami received a fellowship from the National Council for Scientific and Technological Development (CNPq #371497/2013-2). Luisa Matos do Canto received a fellowship from the São Paulo Research Foundation (FAPESP #2014/06323-9 and #2015/25803-4).
CONFLICTS OF INTEREST
The authors declare no competing interests.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study was conducted according to the guidelines of the Declaration of Helsinki, approved by the Human Research Ethics Committee of A.C. Camargo Cancer Center (CEP #2192/16) and the Brazilian National Research Ethics Commission (CONEP #1.721.013). Informed consent was obtained from all subjects involved in the study.
CONSENT FOR PUBLICATION
Not applicable.
AUTHORS’ CONTRIBUTIONS
Conceptualization, S.R.R.; Methodology, C.M.B., L.M.C., A.H.P., and M.M.A.; Formal Analysis, all authors; Investigation, C.M.B., L.M.C., S.S.C., M.N.C.F., and S.A.J.; Resources, S.R.R.; Data Curation, C.M.B., L.M.C., R.A.R.V., and S.S.C.; Writing - Orginal Draft Preparation, L.M.C., R.A.R.V., and S.R.R.; Writing - Review & Editing, all authors; Visualization, C.M.B., L.M.C., and S.R.R.; Supervision, S.R.R. and S.A.J.; Project Administration, S.R.R.; Funding Acquisition, S.R.R. All authors read and approved the final version of the manuscript.
Open Research
DATA AVAILABILITY STATEMENT
The next-generation sequencing data have been deposited at the Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra) with the accession number PRJNA755485.