Serum semaphorin 4C as a diagnostic biomarker in breast cancer: A multicenter retrospective study
Abstract
Background
To date, there is no approved blood-based biomarker for breast cancer detection. Herein, we aimed to assess semaphorin 4C (SEMA4C), a pivotal protein involved in breast cancer progression, as a serum diagnostic biomarker.
Methods
We included 6,213 consecutive inpatients from Tongji Hospital, Qilu Hospital, and Hubei Cancer Hospital. Training cohort and two validation cohorts were introduced for diagnostic exploration and validation. A pan-cancer cohort was used to independently explore the diagnostic potential of SEMA4C among solid tumors. Breast cancer patients who underwent mass excision prior to modified radical mastectomy were also analyzed. We hypothesized that increased pre-treatment serum SEMA4C levels, measured using optimized in-house enzyme-linked immunosorbent assay kits, could detect breast cancer. The endpoints were diagnostic performance, including area under the receiver operating characteristic curve (AUC), sensitivity, and specificity. Post-surgery pathological diagnosis was the reference standard and breast cancer staging followed the TNM classification. There was no restriction on disease stage for eligibilities.
Results
We included 2667 inpatients with breast lesions, 2378 patients with other solid tumors, and 1168 healthy participants. Specifically, 118 patients with breast cancer were diagnosed with stage 0 (5.71%), 620 with stage I (30.00%), 966 with stage II (46.73%), 217 with stage III (10.50%), and 8 with stage IV (0.39%). Patients with breast cancer had significantly higher serum SEMA4C levels than benign breast tumor patients and normal controls (P < 0.001). Elevated serum SEMA4C levels had AUC of 0.920 (95% confidence interval [CI]: 0.900–0.941) and 0.932 (95%CI: 0.911–0.953) for breast cancer detection in the two validation cohorts. The AUCs for detecting early-stage breast cancer (n = 366) and ductal carcinoma in situ (n = 85) were 0.931 (95%CI: 0.916–0.946) and 0.879 (95%CI: 0.832–0.925), respectively. Serum SEMA4C levels significantly decreased after surgery, and the reduction was more striking after modified radical mastectomy, compared with mass excision (P < 0.001). The positive rate of enhanced serum SEMA4C levels was 84.77% for breast cancer and below 20.75% for the other 14 solid tumors.
Conclusions
Serum SEMA4C demonstrated promising potential as a candidate biomarker for breast cancer diagnosis. However, validation in prospective settings and by other study groups is warranted.
Abbreviations
-
- SEMA4C
-
- semaphorin 4C
-
- DCIS
-
- ductal carcinoma in situ
-
- SD
-
- standard deviation
-
- ROC
-
- receiver operating characteristic
-
- AUC
-
- area under the curve
-
- CI
-
- confidence interval
-
- ER
-
- estrogen receptor
-
- PR
-
- progesterone receptor
-
- HER2
-
- human epidermal growth factor receptor 2
-
- IQR
-
- interquartile range
-
- PPV
-
- positive predictive value
-
- NPV
-
- negative predictive value
1 BACKGROUND
Breast cancer remains the leading cause of cancer-related deaths in women and has recently overlapped lung cancer as the leading malignancy worldwide, resulting in nearly 2.3 million new cases and 0.68 million deaths in 2020 [1, 2]. The increasing importance of studying breast cancer could be attributed to the ongoing changes in the risk factors related to societal and economic transitions, including postponement of childbearing, exogenous female hormone use, high body mass index, and physical inactivity [3, 4]. Late-stage presentation is still very common in countries undergoing these transitions, hence, there is an urgent need for early diagnosis of breast cancer.
Currently, breast cancer detection relies on annual/biannual routine mammography screening. For patients with positive mammography results, intensive repeat screening and core-needle biopsy-based histologic diagnosis are performed [5]. As a consequence, these interventions could potentially lead to overtreatment, additional costs, and anxiety. Unlike colon and ovarian cancers, for which carcinoembryonic antigen (CEA) and carbohydrate antigen 125 (CA125) are used for clinical auxiliary diagnosis, there is no effective biomarker for the early detection of breast cancer in the current clinical setting [6, 7].
Identification of effective serological biomarkers with robust sensitivity and specificity, for auxiliary diagnosis of breast cancer, especially early-stage breast cancer is critical for breast cancer diagnosis [8-10]. Numerous biomarkers have been used in previous decades that may aid in the diagnosis of breast cancer, including cancer antigen 153 (CA153), CA125, and CEA. However, low accuracy and reproducibility have impeded their clinical application [11-13]. Thus, the National Comprehensive Cancer Network guidelines still recommend annual mammography screenings for breast cancer surveillance [14]. Recently, serum non-protein biomarkers, such as mutated DNAs, methylated DNAs, and miRNAs, have emerged as a single or panel of diagnostic or prognostic indicators for breast cancer [9, 15–21, 22]. Nevertheless, measurable serum protein biomarkers are still the most applicable candidates for routine clinical assessments and population-based studies because of their favorable properties, including but not limited to less invasive sample collection, low cost, high reproducibility, and operability.
We previously revealed that tumor-associated lymphatic endothelial cells (LECs) upregulated the semaphorin 4C (SEMA4C) expression compared with their normal counterparts using in situ laser capture microdissection of lymphatic vessels and cDNA microarray analysis [23]. Breast cancer-associated LECs not only expressed high levels of membrane-bound SEMA4C but also produced soluble SEMA4C when cleaved by matrix metalloproteinases, which promoted lymphangiogenesis as well as tumor cell migration. Moreover, SEMA4C/PlexinB2 signaling is required for the proliferation of breast cancer cells [24]. Since serum SEMA4C levels were significantly elevated in patients with breast cancer than in normal controls during our preliminary study, we hypothesized that serum SEMA4C might be a diagnostic biomarker for breast cancer. To further investigate this, we performed this large-scale, multicenter, diagnostic study as part of the National Cancer Institute's Early Detection Research Network-Defined Phase 2 Biomarker Study [8]. In this study, we aimed to explore the clinical ability of serum SEMA4C in detecting breast cancer, including early-stage diseases and ductal carcinoma in situ (DCIS).
2 PATIENTS AND METHODS
2.1 Study design
We consecutively included pre-treatment participants including inpatients with benign breast tumors, inpatients with breast cancer, and normal controls to a pre-treatment cohort from Tongji Hospital of Huazhong University of Science and Technology (Wuhan, Hubei, China), Qilu Hospital of Shandong University (Jinan, Shandong, China), and Hubei Cancer Hospital (Wuhan, Hubei, China) between January 1, 2013, and September 30, 2017 (Figure 1). In this study, pre-treatment patients refer to patients who did not undergo any kind of treatment (i.e. surgery, chemotherapy, radiotherapy and target therapy) prior to the blood sample collection, and non-breast cancer controls refer to patients with benign breast lesions and healthy participants. The pre-treatment cohort was divided into a training cohort, validation cohort 1, and validation cohort 2 to determine the optimal cut-off value of serum SEMA4C for breast cancer diagnosis and performance evaluation. The training cohort comprised of patients admitted in Tongji Hospital and Qilu Hospital of Shandong University between January 2013 and June 2016. The validation cohort 1 comprised patients admitted in Tongji Hospital between July 2016 and September 2017. The validation cohort 2 included patients admitted in Hubei Cancer Hospital between July 2016 and June 2017. We also recruited pretreated participants, including patients with breast cancer, patients with other 14 types of solid tumors, and normal controls, to a pan-cancer cohort from 15 cancer centers in Tongji Hospital and Qilu Hospital between July 1, 2017, and June 30, 2018 (Figure 1).
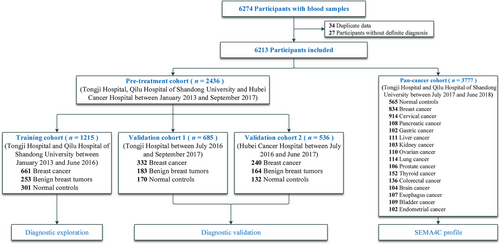
Flow chart of the study design
Abbreviations: SEMA4C, semaphorin 4C
2.2 Study eligibility
Consecutive inpatients with breast lesions and other 14 types of solid tumors, including cervical cancer, pancreatic cancer, gastric cancer, liver cancer, kidney cancer, ovarian cancer, lung cancer, prostate cancer, thyroid cancer, colorectal cancer, brain cancer, esophageal cancer, bladder cancer, and endometrial cancer, from the Surgery Department of the three hospitals were included. The normal controls were volunteers from the Physical Examination Center who were admitted for routine health examinations during that same period. Eligible individuals in the pre-treatment or pan-cancer cohort did not undergo any treatment prior to the blood sample collection. Patients with more than two of the 10 key clinical features missing (age, pathologic diagnosis, histological type, tumor grade, tumor size, lymph node status, metastasis status, estrogen receptor [ER] expression, progesterone receptor [PR] expression, and human epidermal growth factor receptor 2 [HER2] status) and with active inflammatory, autoimmune, renal or liver diseases were excluded. The eligibility criteria also included participants aged over 18 and not having cancer histories other than the investigated ones. Pregnant or lactating women were also excluded from this current study.
Pathological diagnosis after radical surgeries or mass resections was retrieved from the electronic health records for benign breast tumors, breast cancers and other types of solid tumors. Diagnosis was made based on the World Health Organization Classification of Tumors of the Breast Guidelines [25]. The pathologists had no knowledge of the interpretations of patients’ imaging results or serum SEMA4C concentrations, and the pathologic diagnosis was used as the reference standard for diagnosing the patients’ disease. The benign breast lesions included fibroadenoma, intraductal papilloma, atypical ductal hyperplasia, benign phyllodes tumors, lipomas, and hamartomas. Invasive breast cancer and DCIS were categorized as breast cancers. The breast cancer stage was determined according to the Tumor-Node-Metastasis (TNM) classification of the Union for International Cancer Control (7th edition) [26], with stage I (T1N0M0) breast cancer considered as the early stage of the disease.
2.3 Data collection
The clinical information of participants was retrospectively collected from electronic health records by trained investigators after the pathologic diagnosis was obtained and stored in the database of the clinical research center in Tongji Hospital. Data collection was undertaken by two researchers independently (LQ and XL) and supervised by a third investigator (YW).
2.4 Sample collection
The pre-treatment intravenous blood sample was collected into an anticoagulant-free tube (vacuum container) within 48 h prior to treatment. Post-treatment intravenous blood samples were collected 2–8 days after performing breast mass excision or modified radical mastectomy. After centrifugation at 3000 rpm for 10 min, the serum sample was stored at −80°C at each study center. Collection and separation were standardized across centers, with agreed-upon study-specific operating procedures. The serum samples were then shipped on dry ice to the central laboratory of the Cancer Biology Research Center of Tongji Hospital for analysis.
2.5 Chemical materials for enzyme-linked immunosorbent assay (ELISA)
Monoclonal anti-human SEMA4C antibody (#MAB6125, Research & Diagnostics Systems, Incorporated, R&D Systems Inc., Minneapolis, MN, USA) and anti-human SEMA4C antibody (#AF6125, R&D Systems Inc.) were used for analyses. SEMA4C recombinant protein was provided by R&D Systems Inc. (#6125-S4) and was used as the standard protein. The biotin-labeled detection antibody was prepared using a Biotin Labeling Kit-NH2 (Dojindo Molecular Technologies Inc., Kumamoto, Japan).
2.6 Measurement of serum SEMA4C level
Assays for measuring the serum SEMA4C levels were conducted by two researchers (JY and QSC) at the Cancer Biology Research Center of Tongji Hospital who were blinded to the clinical information and pathologic diagnosis of the participants. The serum SEMA4C levels were measured with a double-antibody sandwich ELISA method using in-house SEMA4C detection kits [23]. The ELISA Kit preparations and assays were performed according to the ELISA Development Guide of the R&D Systems Inc. Briefly, 96-well Nunc-immunomicrotiter plates with MaxiSorp surface (Greiner, Germany) were coated with 100 μL of sheep anti-human SEMA4C antibody (4 μg/mL, #AF6125, R&D Systems Inc.) and incubated at 4°C overnight. The reaction was blocked with 1% bovine serum albumin. Sera were diluted by 10-fold and incubated for 2 h at 37°C. The detection antibody, biotinylated mouse anti-human SEMA4C (0.4 μg/mL, #MAB6125, R&D Systems Inc.), was incubated for 2 h at 37°C, followed by the addition of 100 μL at a 1:200 dilution of streptavidin-horseradish peroxidase for 20 min. Color development was achieved by adding 100 μL per well of 3,3,5,5-tetramethylbenzidine and hydrogen peroxide as a substrate, and sulfuric acid (1 mol/L) was added to stop the reaction. The optical density was measured at 450 nm and referenced to 570 nm on a SpectraMax190 plate reader (Molecular Devices, CA, USA). The SEMA4C levels were obtained using linear regression analysis and then fitted to the standard value and multiplied by the dilution factor. When the serum SEMA4C level was <0.1953 ng/mL (the lowest limit of the standard curve), the value was set to zero. All measurements were performed in duplicate. The standard curve covered a range of 0.1953–50.0000 ng/mL. Our in-house SEMA4C ELISA kit was optimized by evaluating the calibration curve, detection limit, recovery, cross-reactivity, dilution linearity. Variations in intra-assay, inter-assay, and day-to-day precision studies were assessed. All measurements were performed in duplicate, and the average values obtained by the two performers were used in the analysis.
2.7 Immunohistochemistry analysis
Specimens from normal/benign-hyperplasic mammary glands (18 cases) and primary invasive breast carcinoma (22 cases) were acquired during surgery as approved by the Ethical Committee of the Medical Faculty of Tongji Medical College (Wuhan, PR China). Tumor specimens were acquired from patients with cancer who had not undergone preoperative radiotherapy or chemotherapy. Tissue sections were subjected to immunohistochemical analysis using the avidin-biotin complex Vectastain Kit (Zsgb-Bio, Beijing, China) according to the manufacturer's protocol. Anti-human CD4 (ab67001, Abcam), anti-human CD8 (ab93278, Abcam), anti-human CD68 (ab213363, Abcam), and anti-human SEMA4C (AF6125, R&D Systems) antibodies were used as primary antibodies. Fixed positive and negative controls were evaluated in each experiment to control the staining variability among batches of experiments. An immunoreactivity-scoring system (HSCORE) was used for the semiquantitative evaluation of protein levels in tissues. Briefly, staining intensity was graded as follows: 0, absence; 1, weak; 2, moderate; 3, strong. The HSCORE score was calculated using the following formula: HSCORE = ∑Pi × i, where i is the staining intensity of immunocytes and Pi is the percentage of corresponding cells at each level of intensity. An HSCORE score of < 1.5 indicated a low protein level while an HSCORE score of ≥ 1.5 indicated a high protein level. Each data point represents the mean score assigned by two pathologists, who were blinded to all clinicopathological variables.
2.8 Statistical analysis
Continuous variables are presented as mean ± standard deviation (SD) or median and interquartile range (IQR), while categorical variables are expressed as absolute frequencies and percentages. The sample size was calculated based on a sensitivity of 80% and a specificity of 90% using the diagnostic test as the present study was not a randomized clinical trial and did not compare other risk factors. The differences in serum SEMA4C values and other continuous variables between the two independent groups were tested using the Mann-Whitney U test. Pearson's chi-square test or Fisher's exact test was used to analyze the association between pre-treatment serum SEMA4C levels and clinical or pathological factors. A receiver operating characteristic (ROC) curve was developed to determine the area under the curve (AUC). The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated with their corresponding 95% confidence intervals (CIs). PPV is a diagnostic statistic that detects the presence of diseases and is calculated as follows: PPV = true positive cases/(true positive cases + false-positive cases). In contrast, NPV is a diagnostic statistic that detects the absence of diseases and is calculated as follows: NPV = true negative cases/(true negative cases + false-negative cases). The optimal cut-off value for the training cohort was obtained by maximizing the sum of sensitivity and specificity, minimizing the overall error [square root of the sum of (1−sensitivity)2 + (1–specificity)2], and minimizing the distance of the cut-off value to the top-left corner of the ROC curve [27]. All statistical analyses were performed using the Statistical Package for the Social Sciences (SPSS v23.0, IBM Corp., NY, USA) and R software (v3.4.1; www.r-project.org). Statistical significance was set at P < 0.05.
3 RESULTS
3.1 Baseline characteristics of the study participants
Overall, 6213 participants were consecutively included (Figure 1): 1215 pre-treatment patients were assigned to the training cohort, 685 pre-treatment patients were assigned to the validation cohort 1, 536 pre-treatment patients were assigned to the validation cohort 2, and 3777 pretreatment cases were assigned to the pan-cancer cohort, which included healthy participants, patients with breast cancer, and those with other 14 types of solid tumors.
The demographic and clinicopathological characteristics of the training cohort, validation cohort 1, and validation cohort 2 are summarized in Table 1. The overall frequencies at diagnosis of stages I, II, and III were 34.49%, 47.20%, and 9.38% in the training cohort; 22.59%, 38.86%, and 10.84% in validation cohort 1; and 24.58%, 50.83%, and 11.67% in validation cohort 2, respectively. Only four patients diagnosed with stage IV disease were included, two from the training cohort and two from the validation cohort 1. The training cohort and validation cohorts were matched for the mean age between breast cancer patients and normal controls; patients with benign breast tumors were younger than those with cancerous tumors as benign breast tumors were observed to develop at a younger age (P < 0.05).
| Characteristic | Training Cohort | Validation Cohort 1 | Validation Cohort 2 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Normal controls n (%) | Benign breast tumors n (%) | Breast cancer n (%) | Normal controls n (%) | Benign breast tumors n (%) | Breast cancer n (%) | Normal controls n (%) | Benign breast tumors n (%) | Breast cancer n (%) | |
| Age, years | |||||||||
| Mean (SD) | 49.50 (12.50) | 40.11 (11.58) | 50.20 (10.48) | 46.39 (14.22) | 40.74 (11.34) | 48.31 (10.34) | 47.33 (12.35) | 41.15 (9.64) | 51.20 (9.67) |
| <35 | 44 (14.62%) | 79 (31.22%) | 36 (5.45%) | 32 (18.82%) | 58 (31.69%) | 33 (9.94%) | 15 (11.36%) | 38 (23.17%) | 9 (3.75%) |
| 35–49 | 71 (23.59%) | 130 (51.38%) | 303 (45.84%) | 68 (40.00%) | 90 (49.18%) | 157 (47.29%) | 54 (40.91%) | 94 (57.32%) | 96 (40.00%) |
| 50–70 | 180 (59.80%) | 43 (17.00%) | 295 (44.63%) | 61 (35.88%) | 35 (19.13%) | 134 (40.36%) | 60 (45.45%) | 32 (19.51%) | 109 (45.42%) |
| >70 | 6 (1.99%) | 1 (0.40%) | 26 (3.93%) | 9 (5.29%) | 0 (0) | 8 (2.41%) | 3 (2.27%) | 0 (0) | 8 (3.33%) |
| Missing, n | 0 (0) | 0 (0) | 1 (0.15%) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 18 (7.50%) |
| Histological type | |||||||||
| DCIS | 23 (3.48%) | 62 (18.67%) | 0 (0) | ||||||
| IDC | 607 (91.83%) | 237 (71.39%) | 220 (91.67%) | ||||||
| ILC | 20 (3.03%) | 0 (0) | 3 (1.25%) | ||||||
| Missing, n | 11 (1.66%) | 33 (9.94%) | 17 (7.08%) | ||||||
| Tumor grade | |||||||||
| Grade I-II | 478 (72.31%) | 232 (69.88%) | 166 (69.17%) | ||||||
| Grade III | 145 (21.94%) | 65 (19.58%) | 51 (21.25%) | ||||||
| Missing, n | 38 (5.75%) | 35 (10.54%) | 23 (9.58%) | ||||||
| Tumor size, cm | |||||||||
| ≤2 | 287 (43.42%) | 160 (48.19%) | 92 (38.33%) | ||||||
| 2<, ≤5 | 312 (47.2%) | 143 (43.07%) | 104 (43.33%) | ||||||
| >5 | 26 (3.93%) | 17 (5.12%) | 15 (6.25%) | ||||||
| Missing, n | 36 (5.45%) | 12 (3.61%) | 29 (12.08%) | ||||||
| Pathological lymph node status | |||||||||
| pN0 | 502 (75.95%) | 177 (53.31%) | 111 (46.25%) | ||||||
| pN1 | 72 (10.89%) | 63 (18.98%) | 66 (27.5%) | ||||||
| pN2 | 33 (4.99%) | 18 (5.42%) | 0 (0) | ||||||
| pN3 | 24 (3.63%) | 18 (5.42%) | 0 (0) | ||||||
| pNx | 6 (0.91%) | 23 (6.93%) | 46 (19.17%) | ||||||
| Missing, n | 24 (3.63%) | 33 (9.94%) | 17 (7.08%) | ||||||
| Metastasis status | |||||||||
| M0 | 658 (99.55%) | 310 (93.37%) | 240 (100%) | ||||||
| M1 | 2 (0.30%) | 4 (1.20%) | 0 (0) | ||||||
| Missing, n | 1 (0.15%) | 18 (5.42%) | 0 (0) | ||||||
| TNM stage | |||||||||
| I | 228 (34.49%) | 75 (22.59%) | 59 (24.58%) | ||||||
| II | 312 (47.20%) | 129 (38.86%) | 122 (50.83%) | ||||||
| III | 62 (9.38%) | 36 (10.84%) | 28 (11.67%) | ||||||
| IV | 2 (0.30%) | 2 (0.60%) | 0 (0) | ||||||
| Missing, n | 34 (5.14%) | 28 (8.43%) | 31 (12.92%) | ||||||
| Hormone receptor status | |||||||||
| ER+, PR+, or both | 466 (70.50%) | 220 (66.27%) | 151 (62.92%) | ||||||
| ER– and PR− | 187 (28.29%) | 88 (26.51%) | 78 (32.50%) | ||||||
| Missing, n | 8 (1.21%) | 24 (7.23%) | 11 (4.58%) | ||||||
| HER2 status | |||||||||
| Negative | 308 (46.6%) | 202 (60.84%) | 109 (45.42%) | ||||||
| Positive | 328 (49.62%) | 103 (31.02%) | 106 (44.17%) | ||||||
| Missing, n | 25 (3.78%) | 27 (8.13%) | 25 (10.42%) | ||||||
| Clinicopathologic subtype | |||||||||
| HER2−: ER+, PR+, or both | 237 (35.85%) | 146 (43.98%) | 72 (30.00%) | ||||||
| HER2+: ER+, PR+, or both | 212 (32.07%) | 60 (18.07%) | 71 (29.58%) | ||||||
| HER2+: ER−, PR− | 116 (17.55%) | 43 (12.95%) | 35 (14.58%) | ||||||
| Triple-negative | 65 (9.83%) | 41 (12.35%) | 37 (15.42%) | ||||||
| Missing, n | 31 (4.69%) | 42 (12.65%) | 25 (10.42%) | ||||||
- Ages are presented as mean ± standard deviation.
- Categorical data were summarized as absolute frequencies and percentages.
- Abbreviations: DCIS, ductal carcinoma in situ. IDC, invasive ductal carcinoma. ILC, invasive lobular carcinoma. ER, estrogen receptor. PR, progesterone receptor. HER2, human epidermal growth factor receptor 2.
3.2 Diagnostic value of serum SEMA4C in breast cancer
To accurately measure the serum SEMA4C, we optimized the in-house SEMA4C ELISA detection system by evaluating the calibration curve, detection limit, recovery, cross-reactivity, and dilution linearity (Supplementary Figure S1). The variation in intra-assay, inter-assay, and day-to-day precision studies was limited to less than 10% (Supplementary Table S1).
In the training cohort, the serum SEMA4C levels were significantly higher in patients with breast cancer (7.46 ± 2.69 ng/mL) than in those with benign breast tumors (3.51 ± 1.60 ng/mL) and normal controls (3.19 ± 0.96 ng/mL; both P < 0.001) (Figure 2A, Supplementary Table S2). Meanwhile, the serum SEMA4C levels were not significantly different between patients with benign breast lesions and normal controls (P < 0.05). The optimal cut-off value of serum SEMA4C for diagnosing breast cancer was 5.00 ng/mL when an adjacent integer was chosen from the optimal cut-off values of 5.125 ng/mL in Tongji Hospital and 4.754 ng/mL in Qilu Hospital (Supplementary Table S3). The AUC was 0.938 (95% CI: 0.924–0.951), with a sensitivity of 84.4% and a specificity of 89.9% (Figure 2B and Table 2).
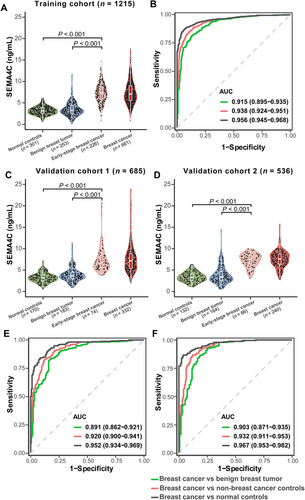
Diagnostic value of SEMA4C in breast cancer
In the training cohort (A), validation cohort 1 (C), and validation cohort 2 (D), the pre-treatment serum SEMA4C level was significantly higher in patients with breast cancer including those with early-stage disease than in normal controls and those with benign breast tumors. For (A), (C), and (D), the distribution of serum SEMA4C levels represent as violin plots showing the frequency (width of density plot), median (white dot), interquartile range (bar), and 95% CI (line). In the training cohort (B), validation cohort 1 (E), and validation cohort 2 (F), elevated serum SEMA4C levels had high AUCs in diagnosing breast cancer. AUC values are presented with 95% confidence interval. The differences in serum SEMA4C values between two independent groups were tested using the Mann-Whitney U test. Breast cancer included Early-stage breast cancer.
Abbreviations: BC, breast cancer. SEMA4C, semaphorin 4C. AUC, area under the receiver operating characteristic curve
| Characteristic | No. of patients | AUC (95% CI) | SN (%) | SP (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|
| Training cohort | ||||||
| Breast cancer vs non-breast cancer controls | 661 vs 554 | 0.938 (0.924–0.951) | 84.4 | 89.9 | 90.9 | 82.9 |
| Early-stage breast cancer vs non-breast cancer controls | 226 vs 554 | 0.936 (0.916–0.957) | 84.1 | 91.5 | 80.2 | 93.4 |
| Breast cancer vs benign breast tumor | 661 vs 253 | 0.915 (0.895–0.935) | 81.2 | 87.4 | 94.4 | 64.1 |
| Breast cancer vs normal controls | 661 vs 301 | 0.956 (0.945–0.968) | 86.7 | 94.0 | 97.0 | 76.3 |
| Validation cohort 1 | ||||||
| Breast cancer vs non-breast cancer controls | 332 vs 353 | 0.920 (0.900–0.941) | 82.8 | 87.5 | 86.2 | 84.4 |
| Early-stage breast cancer vs non-breast cancer controls | 74 vs 353 | 0.937 (0.913–0.960) | 90.5 | 86.7 | 58.8 | 97.8 |
| Breast cancer vs benign breast tumor | 332 vs 183 | 0.891 (0.862–0.921) | 82.8 | 83.1 | 89.9 | 72.7 |
| Breast cancer vs normal controls | 332 vs 170 | 0.952 (0.934–0.969) | 84.0 | 91.8 | 95.2 | 74.6 |
| Validation cohort 2 | ||||||
| Breast cancer vs non-breast cancer controls | 240 vs 296 | 0.932 (0.911–0.953) | 86.7 | 87.8 | 85.2 | 89.0 |
| Early-stage breast cancer vs non-breast cancer controls | 66 vs 296 | 0.929 (0.899–0.958) | 86.4 | 87.8 | 61.3 | 96.7 |
| Breast cancer vs benign breast tumor | 240 vs 164 | 0.903 (0.871–0.935) | 86.7 | 81.1 | 87.0 | 80.6 |
| Breast cancer vs normal controls | 240 vs 132 | 0.967 (0.953–0.982) | 86.7 | 96.2 | 97.7 | 79.9 |
| Total | ||||||
| Early-stage breast cancer vs non-breast cancer controls | 366 vs 1203 | 0.931 (0.916–0.946) | 85.2 | 88.9 | 70.0 | 95.2 |
| DCIS vs non-breast cancer controls | 85 vs 1203 | 0.879 (0.832–0.925) | 74.1 | 89.7 | 33.7 | 98.0 |
| DCIS vs benign breast tumor | 85 vs 600 | 0.855 (0.806–0.904) | 74.1 | 84.2 | 39.9 | 95.8 |
| DCIS vs normal controls | 85 vs 603 | 0.902 (0.856–0.948) | 74.1 | 95.2 | 68.5 | 96.3 |
- Abbreviations: AUC, area under the receiver operating characteristic curve. CI, confidence interval. DCIS, ductal carcinoma in situ. SN, sensitivity. SP, specificity. NPV, negative predictive value. PPV, positive predictive value. Breast cancer, invasive breast cancer and DCIS. Non-breast cancer controls, normal controls and patients with breast benign tumors. Early-stage breast cancer, T1N0M0 invasive breast cancer.
Similarly, the serum SEMA4C levels in the two validation cohorts were significantly higher in patients with breast cancer than in those with benign breast tumors and normal controls (both P < 0.001; Figure 2C and D and Supplementary Table S2), and the difference between benign tumors and normal controls was not significant (P > 0.05). Based on the determined optimal threshold in the training cohort, serum SEMA4C showed an AUC value of 0.920 (95% CI: 0.900–0.941), a sensitivity of 82.8%, and a specificity of 87.5% for the detection of breast cancer in the validation cohort 1 (Figure 2E and Table 2). For validation cohort 2, the AUC was 0.932 (95% CI: 0.911–0.953), with a sensitivity of 86.7% and a specificity of 87.8% (Figure 2F and Table 2).
In addition, the pre-treatment serum SEMA4C levels were not associated with most clinicopathological features in the training and validation cohorts, including tumor size, tumor grade, lymph node status, and ER/PR/HER2 status (Supplementary Table S4).
3.3 Diagnostic value of serum SEMA4C in early-stage disease and DCIS
Late-stage breast cancer is still prevalent in many countries. We examined whether increased serum SEMA4C levels could be used to identify early-stage breast cancer, considering that there is no available effective blood-based biomarker for the early detection of this disease. Table 2 shows that early-stage breast cancer could be accurately distinguished from non-breast cancer controls with high AUCs (training cohort, 0.936 [95% CI: 0.916–0.957]; validation cohort 1, 0.937 [95% CI: 0.913–0.960]; validation cohort 2, 0.929 [95% CI: 0.899–0.958]). DCIS accounts for 15% of all breast cancer types; however, no serum protein marker has been validated for DCIS diagnosis or screening [28]. In this present study, DCIS was diagnosed in 85 (6.89%) inpatients with breast cancer. When differentiating DCIS patients from non-breast cancer controls; serum SEMA4C yielded an AUC of 0.879 (95% CI: 0.832–0.925), sensitivity of 74.1%, and specificity of 89.7% (Supplementary Figure S2, Table 2, and Supplementary Table S5). Briefly, increased serum SEMA4C levels exhibited high accuracy in discriminating early-stage breast cancers/DCIS from non-breast cancer controls.
3.4 Measurements of serum SEMA4C in patients with breast cancer before and after surgery
To investigate whether serum SEMA4C could help measure the response to surgery in patients with breast cancer, we compared the SEMA4C levels before treatment with those after breast mass excision or modified radical mastectomy in paired samples. For the 140 patients who underwent modified radical mastectomy, the post-surgery serum SEMA4C level was significantly lower than the pre-treatment level (P < 0.001; Figure 3A, Supplementary Tables S6 and S7). They had undergone breast mass excision prior to modified radical mastectomy as they were misdiagnosed with benign breast lesions pre- or intraoperatively. Notably, the SEMA4C level after breast mass excision of these patients (6.48 ± 3.11 ng/mL) was significantly higher than that after modified radical mastectomy (5.26 ± 3.00 ng/mL) (P < 0.001; Figure 3A and Supplementary Table S7).
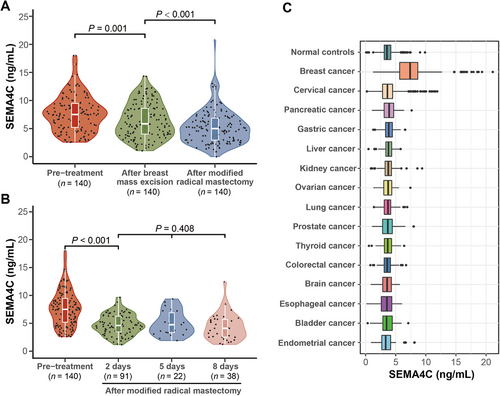
Measurements of serum SEMA4C in patients with breast cancer before and after surgery and in patients with different types of solid cancers and normal controls
(A) The serum SEMA4C levels of patients who underwent modified radical mastectomy were lower than those of patients who underwent mass excision. (B) By measuring the serum SEMA4C level at different days (2 days, 5 days, and 8 days) after surgery, we found that the serum SEMA4C levels decreased with time after surgery. (C) Serum SEMA4C levels were significantly higher in patients with breast cancer than in those with other 14 types of solid tumors and normal controls.
Abbreviations: SEMA4C, semaphorin 4C
Meanwhile, 151 serum samples were collected from patients with breast cancer after undergoing modified radical mastectomy: 91 samples at day-2, 22 at day-5, and 38 at day-8. The mean serum SEMA4C level before surgery was 7.75 ± 3.25 ng/mL, and the values declined afterward (4.80 ± 1.85 ng/mL at day-2; 5.01 ± 2.19 ng/mL at day-5; and 4.36 ± 2.34 ng/mL at day-8; Figure 3B, Supplementary Table S7).
Generally, decreased serum SEMA4C levels were observed after modified radical mastectomy or breast mass excision, showing the potential of serum SEMA4C in assessing the response to surgery in breast cancer patients.
3.5 Measurements of serum SEMA4C in patients with solid tumors of 15 origins and healthy controls
To further explore whether SEMA4C could be used as a biomarker specific to breast cancer among common solid tumor patients, a pan-cancer cohort was established. The results showed that serum SEMA4C levels were significantly higher in patients with breast cancer than in those with other 14 types of solid tumors and normal controls (P < 0.001; Figure 3C and Supplementary Tables S8–S10). Compared with the normal controls, the serum SEMA4C levels were slightly higher in patients with cervical, pancreatic, gastric, liver, kidney, and ovarian cancers (P > 0.05), and slightly lower in patients with lung, prostate, thyroid, colorectal, brain, esophageal, bladder or endometrial cancers (P > 0.05). Based on a cut-off value of 5.00 ng/mL for serum SEMA4C levels, 84.77% (707/834), 15.75% (144/914), 19.44% (21/108), 12.75% (13/102), 12.61% (14/111), 12.62% (13/103), 9.09% (10/110), 9.65% (11/114), 20.75% (22/106), 7.89% (12/152), 8.08% (11/136), 8.65% 9(/104), 8.41% (9/107), 8.26% (9/109), and 7.84% (8/102) of the patients tested positive for breast, cervical, pancreatic, gastric, liver, kidney, ovarian, lung, prostate, thyroid, colorectal, brain, esophageal, bladder, and endometrial cancers, respectively. Generally, despite the differences in age distribution among female patients, high serum SEMA4C levels were specifically observed among patients with breast cancer (Supplementary Table S11).
4 DISCUSSION
This study aimed to evaluate the utility and robustness of serum SEMA4C as a diagnostic biomarker for breast cancer. We demonstrated that measurements of serum SEMA4C levels before treatment (i.e. surgery, chemotherapy, radiotherapy and targeted therapy) enabled the accurate discrimination between patients with breast cancer, including those with early-stage diseases and DCIS, and patients with benign breast tumors or normal controls. Serum SEMA4C levels significantly decreased after surgery, and the reduction was more striking after modified radical mastectomy, compared with mass excision (P < 0.001). In addition, high serum SEMA4C levels were specifically observed in patients with breast cancer of stage 0 to IV.
An ideal serological protein biomarker should meet at least two criteria. First, it should be a secreted protein that can be easily detected in the serum. Second, it should be specifically overexpressed in cancer rather than in normal tissues or benign lesions [8]. SEMA4C, a transmembrane group-4 semaphorin, is predominantly expressed in the neuronal tissue of embryos. It was originally identified as a regulator of axon growth during the development of the central nervous system [29, 30]. We previously identified that SEMA4C was differentially expressed in normal lymphatic vessels and their breast cancer-associated counterparts, and SEMA4C was cleaved by matrix metalloproteinases to release a soluble form [23]. Therefore, SEMA4C meets the following two criteria: it is a secretory protein that is specifically overexpressed in tumor-associated lymphatic vessels and is barely detectable in normal adult tissues.
We measured the serum SEMA4C levels in patients with breast cancer, patients with benign breast tumors, normal controls, and patients with other 14 types of solid tumors, which indicated that serum SEMA4C level is a biomarker specific for breast cancer diagnosis. With an optimal SEMA4C cut-off value of 5.00 ng/mL, the results suggested that the serum SEMA4C was a specific biomarker of breast cancer and clearly discriminated patients with breast cancer from non-breast cancer controls in the training cohort and two independent validation cohorts. Diagnosing breast cancer using radio-free and noninvasive serum biomarkers has attracted broad attention among researchers. Recent studies have focused on the use of transcriptomic or epigenetic panels for breast cancer diagnosis. In 2020, Zou et al. [31] developed a 12-miRNA panel, which yielded an AUC of 0.94, a sensitivity of 0.84, a specificity of 0.91, a PPV of 0.90, and an NPV of 0.85, in 216 patients with breast cancer and 214 normal controls. Several other lncRNAs (AUC 0.74–0.95) [32-34], microRNA (AUC 0.86–0.99) [35-37], and DNA methylation panels (AUC 0.58–0.98) [38] also showed high diagnostic accuracy, which is corresponded to the accuracy of serum SEMA4C levels in diagnosing breast cancer. However, these tests are not always readily available and lack clinical application in low-resource settings. Clinically accessible proteinic biomarkers are cheaper but have relatively low accuracy. CA153 and CEA are the most frequently evaluated serum biomarkers for breast cancer detection. However, only 50% of breast cancers are detected based on the CA153 or CEA levels [39]. The sensitivity, specificity, PPV, and NPV of CA153 and CEA (52.8%, 61.4%, 56.1%, and 56.8%; 69.1%, 49.3%, 62.5%, and 51.2%, respectively) were lower than those of the serum SEMA4C levels. Recent studies have also revealed the diagnostic potential of several new biomarkers, including CPT1A [40], CPN1 [41], and others [42, 43]; however, the reliability of their research is limited due to the small sample size. In this study, serum SEMA4C levels were observed to have a higher AUC (between 0.920 and 0.938), with a sensitivity of 84.4% and a specificity of 89.9%, a PPV of 90.9%, and an NPV of 82.9%. However, the current results are insufficient to support the accuracy of SEMA4C levels for detecting breast cancer in clinical practice, and more studies are warranted to validate its diagnostic performance and explore its potential when integrated into the current breast cancer diagnostic processes. For example, the combination of serum SEM4AC level and breast imaging could achieve better accuracy for diagnosing breast cancers. Notably, the potential diagnostic value of SEMA4C was also observed in patients with breast cancer in situ and with clinical stage T1N0M0. It would be clinically valuable to provide timely treatment if SEMA4C could detect breast cancer at an early stage.
Interestingly, pre-treatment serum SEMA4C levels were not correlated with most clinicopathological characteristics of breast cancer, including tumor size, tumor grade, lymph node status, and ER/PR/HER2 status, which is uncommon for cancer biomarkers. Thus, we hypothesized that the factors influencing serum SEMA4C levels lie in the non-cancerous components within the tumor microenvironment, not in the tumor cells. One supporting evidence is that we observed a significant difference in mean serum SEMA4C levels between breast cancer patients who underwent breast mass excision (6.48 ± 3.11 ng/mL) and those who underwent modified radical mastectomy (5.26 ± 3.00 ng/mL; P < 0.001) (Figure 3A and Supplementary Table S7). This disparity may be attributed to the removal of both tumor mass and peripheral tumoral tissue during a modified radical mastectomy, in contrast with only the removal of the primary tumor during a breast mass excision. Moreover, SEMA4C was first identified in tumor-associated lymphatic vessels and was mainly overexpressed in peripheral tumoral LECs and immunocytes (Supplementary Figure S3), but was weakly expressed in tumor tissues. Increased serum SEMA4C levels might be a result of enhanced expression or secretion of SEMA4C from non-breast cancer cells within the tumor microenvironment. This could explain why serum SEMA4C levels significantly increased in patients with DCIS and early-stage breast cancer because the tumor interstitium frequently changes prior to tumorigenesis. However, this hypothesis should be thoroughly investigated in future studies.
This study had several limitations. First, it is retrospective and tumor tissue sections were not available to assess the SEMA4C expression in the tissue of solid tumors. The interpretation of results should be further validated in a prospective setting. Second, as all study participants were Chinese, the accuracy of serum SEMA4C in diagnosing breast cancer must be validated in other populations. Third, the influence of common factors, including obesity, smoking, and alcohol consumption, was not measured in the current study and could be associated with potential bias. Fourth, the number of patients with DCIS and other solid tumors was relatively small. A larger sample size is necessary to obtain more solid conclusions.
5 CONCLUSIONS
To the best of our knowledge, this is the first large-scale, multicenter study to report the diagnostic performance of serum SEMA4C levels in breast cancer. Our findings, together with the existing evidence, highlight the diagnostic value of serum SEMA4C in discriminating breast cancers (including DCIS and early-stage disease) from non-breast cancer diseases. For clinical implementation, further prospective study studies are needed.
ACKNOWLEDGMENTS
This work was supported by the National Science and Technology Major Sub-Project (2018ZX10301402-002), the National Natural Science Foundation of China (81772787, 81902653, and 82072889), the Technical Innovation Special Project of Hubei Province (2018ACA138), the Fundamental Research Funds for the Central Universities (2019kfyXMBZ024), and Municipal Health Commission Project of Wuhan (WX18Q16). Special thanks to Lu Y for technical support and to Yidu Cloud (Beijing) Technology Co., Ltd. and W. Wood for revising this manuscript.
AUTHORS’ CONTRIBUTIONS
YW and LQ designed the study, performed the experiments, and analyzed and interpreted the data. XL and JHL analyzed and interpreted the data. JY and SQC performed the ELISA. YQD read the slides. JY and JXW established an ELISA. HYL, DL, FY, ESG, TF, XTL, JJM, PJ, JCW, MFW, QX, QZ, JBH, ZYY, GC, XYW, and QHZ provided, analyzed, and interpreted the patient samples and clinical data. LZ, YX, YQW, JS, YJF, XRL, and SXW provided the patient samples, clinical data, or both. Mona PT, Kazuaki T, QFY, and BHK provided advice on the conception and design of the study. QLG and DM conceptualized and designed the study, supervised the project, analyzed and interpreted the data, and wrote the paper. All authors are responsible for the respective data and analysis, approved the final version of the manuscript, and agreed to the submission of this manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the ethical committee of Tongji Hospital, Qilu Hospital, and Hubei Cancer Hospital (TJ-C20140311). All participants provided written informed consent prior to their inclusion in the study.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author (Qinglei Gao, [email protected] ) upon reasonable request.




