CircBA1 derived from BCR-ABL fusion gene inhibits cell proliferation in chronic myeloid leukemia
Abbreviations
-
- CML
-
- chronic myeloid leukemia
-
- FISH
-
- fluorescence in situ hybridization
-
- PBS
-
- phosphate buffer saline
-
- PCR
-
- polymerase chain reaction
-
- qRT-PCR
-
- quantitative reverse transcription PCR
-
- RT-PCR
-
- reverse transcription PCR
-
- shRNAs
-
- short hairpin RNAs
Dear editor,
Chronic myeloid leukemia (CML) is a malignant myeloproliferative disorder arising from hematopoietic stem cells and accounts for 15% of newly diagnosed cases of leukemia in adults. Central to the pathogenesis of CML is the presence of the BCR-ABL fusion gene resulting from the chromosome translocation t (9;22) (q34; q11) [1]. The chimeric oncoprotein encoded by BCR-ABL exhibits aberrant tyrosine kinase activity and drives oncogenic processes by dysregulating or reprogramming multiple downstream signaling pathways involved in cell proliferation, differentiation, and survival [2]. Recently, emerging evidences have demonstrated that fusion genes can not only encode oncoprotein but also generate circular RNAs (circRNAs) involved in tumor progression [3].
CircRNAs are a kind of non-coding RNAs with a covalently closed loop structure that confers them not to be digested by exonuclease [4]. Mounting evidence have established circRNAs as playing indispensable oncogenic or tumor-suppressive roles in various tumors, including CML [5-8]. For instance, upregulated circHIPK3 exerts an oncogenic role in CML and is an indicator of poor prognosis in CML patients [5]. Circ_0009910 and circ_100053 have also been shown to be associated with poor prognosis and imatinib resistance [6, 7]. A fusion circRNA (circBA9.3) derived from the BCR-ABL fusion gene was reported to promote cell proliferation of CML by upregulating c-ABL1 or BCR-ABL1 protein levels [8]. Due to the crucial role of the BCR-ABL fusion gene in CML, it is worth exploring whether the BCR-ABL fusion gene could produce other functional fusion circRNAs involved in CML progression. In this study, we aimed to identify the existence of fusion circRNA (circBA1) generated from the BCR-ABL fusion gene in BCR-ABL positive CML cell lines, and explore the crucial role of circBA1 in CML.
The methods of this study are provided in the Supplementary information file. Briefly, two CML cell lines (K562 and KU812) were used for this study as they possess the BCR-ABL fusion gene. Reverse transcription polymerase chain reaction (RT-PCR) and Sanger sequencing conducted by convergent primer (F1/R1) confirmed the fusion site of e14a2 BCR-ABL in these two cell lines (Figure 1A and Supplementary Fig. S1A). Next, total RNAs from K562 and KU812 cells were subjected to nested PCR by using two pairs of the convergent primers (F2/R2 and F3/R3) to identify the fusion circRNAs. As shown in Figure 1B and Supplementary Fig. S1B, a 635 bp product was detected in both K562 and KU812 cells. Sanger sequencing revealed that the product contained the back-splicing junction site between BCR exon 9 and ABL exon 3, indicating that the BCR-ABL fusion gene can produce a circular RNA (named circBA1), containing complete BCR exons 10-14 and ABL exon 2, as well as a partial sequence of BCR exon 9 and ABL exon 3 (Figure 1C). Results from CML patient samples confirmed the existence of circBA1 (Figure 1D and 1E).
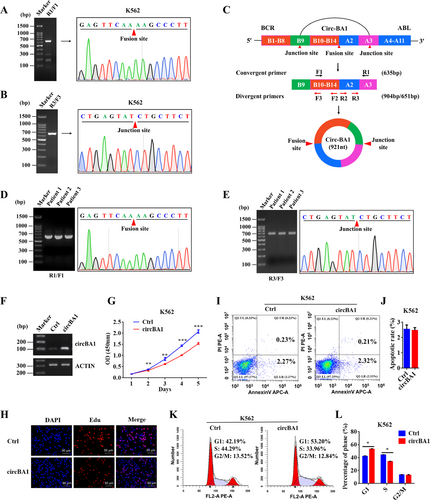
CircBA1 derived from the BCR-ABL fusion gene inhibits the cell proliferation of K562 cells. (A) Identification of the fusion site between BCR and ABL genes in K562 cells using the convergent primers F1/R1. (B) Identification of the fusion circBA1 in K562 cells using nested PCR and Sanger sequencing. (C) Schematic representation of circBA1. B9 represents BCR exon 9; A3 represents ABL exon 3. (D) Detection of the BCR-ABL fusion type in CML patients. (E) Identification of circBA1 in CML patients using nested PCR and Sanger sequencing. (F) Identification of circBA1 overexpression by semi-quantitative PCR in K562 cells. (G, H) CCK-8 assay (G) and Edu assay (H) of K562 cells stably overexpressing circBA1. (I, J) Effect of circBA1 overexpression on cell apoptosis in K562 cells. (K, L) Cell cycle analysis of K562 cells stably overexpressing circBA1. Abbreviations: CCK-8, Cell counting kit-8; OD, Optical density; Edu, 5-Ethynyl-2′-deoxyuridine; PI, Propidium iodide; Ctrl, Control
To investigate the circular characteristics of circBA1, total RNAs were digested with RNase R to remove linear RNAs, and then subjected to RT-PCR with the specific primer covering the circBA1 junction site. The results showed that circBA1 was resistant to RNase R digestion compared to BCR-ABL linear mRNA, indicating that circBA1 was circular (Supplementary Fig. S2A). Analyzing the stability of circBA1 and BCR-ABL linear mRNA in K562 and KU812 cell lines treated with actinomycin D, an inhibitor of transcription, revealed that circBA1 was much more stable than the linear RNAs in both cells, as the half-life of circBA1 exceeded 24h (Supplementary Fig. S2B). To observe the cellular localization of circBA1, we performed subcellular fractionation analyses and found that circBA1 was preferentially located in the cytoplasm of both K562 and KU812 cells (Supplementary Fig S2C). Fluorescence in situ hybridization (FISH) using the probe crossing the junction site further confirmed the cytoplasmic distribution of circBA1 (Supplementary Fig. S2D).
To explore the biological function of circBA1 in CML, we ectopically expressed circBA1 in a special vector containing upstream and downstream flanking repeat sequences which facilitate the formation of circRNA (Supplementary Fig. S3A). Then, the upstream and downstream flanking sequences together with circBA1 were removed to a lentiviral vector to achieve stable overexpression of circBA1 in K562 and KU812 cells (Supplementary Fig. S3B). Semi-quantitative PCR results showed that circBA1 was successfully overexpressed in K562 (Figure 1F) and KU812 (Supplementary Fig. S1C) cells. RT-PCR and Sanger sequencing were subsequently used to confirm the junction site of overexpressed circBA1 in K562 and KU812 cells (Supplementary Fig. S3C and S3D). Cell proliferation assay revealed that overexpression of circBA1 significantly suppressed CML cell growth (Figure 1G and Supplementary Fig. S1D), which were further confirmed by Edu assays (Figure 1H and Supplementary Fig. S1E). However, cell apoptosis assay showed that circBA1 had little effect on CML cell apoptosis (Figure 1I and 1J, Supplementary Fig. S1F and S1G). To understand whether the inhibition of CML cell proliferation induced by circBA1 was associated with cell cycle changes, we analyzed the cell cycle distribution of K562 and KU812 cells. Our results illustrated that the overexpression of circBA1 increased the number of cells in the G1 phase, with a corresponding reduction in the number of cells in the S phase; suggesting that circBA1 induced cell cycle arrest in CML. This phenomenon was observed in both K562 (Figure 1K and 1L) and KU812 (Supplementary Fig. S1H and S1I) cells.
Besides gain-of-function experiments, we also implemented a loss-of-function strategy to examine the biological function of circBA1. Short hairpin RNAs (shRNAs) specifically targeting the back-splicing junction of circBA1 (Supplementary Fig. S3E and S3F) were designed to efficiently knockdown circBA1 expression in K562 (Supplementary Fig. S4A) and KU812 (Supplementary Fig. S4D) cells. CCK-8 and Edu assays revealed that the knockdown of circBA1 significantly promoted CML cell proliferation (Supplementary Fig. S4B, S4C, S4E, and S4F). Silencing circBA1 had little influence on cell apoptosis in both K562 (Supplementary Fig. S4G) and KU812 (Supplementary Fig. S4H) cells. Further, the knockdown of circBA1 promoted cell cycle progression by decreasing the number of cells in G1 phase and increasing the number of cells in S phase (Supplementary Fig. S4I and S4J).
Fusion circRNAs generated from oncogenic fusion genes were identified to play curial roles in tumor progression [3], but little is known about fusion circRNAs derived from the BCR-ABL fusion gene. In this present study, we successfully identified a fusion circRNA (circBA1) derived from the BCR-ABL fusion gene in CML cell lines and patient samples, and found that circBA1 could inhibit the cell proliferation of CML cells by inducing cell cycle arrest. A previous study demonstrated that fusion circBA9.3 was involved in tyrosine-kinase inhibitors (TKIs) resistance by reducing TKIs induced cell apoptosis [8]. However, in our study, we did not treat CML cells with TKIs, and we found that circBA1 had little effect on apoptosis of CML cells. Recent studies have demonstrated that circRNAs played important roles in cell proliferation and transformation of leukemia [7-9]. Some reported circRNAs in leukemia were upregulated and exerted an oncogenic role [7, 8], while others acted as a tumor suppressor. CircFBXW7, a well-established tumor suppressor in glioma, also plays a tumor-suppressive role in acute myeloid leukemia (AML). A recent study demonstrated that high expression of circFBXW7 was associated with positive regulation of leukocyte differentiation while depletion of circFBXW7 promoted cell proliferation of AML [9]. In this study, we highlighted the tumor-suppressive role of fusion circBA1 in CML. Our findings revealed that the BCR-ABL translocation could generate a tumor-suppressive fusion circRNA, expanding the current knowledge regarding oncogenic fusion genes and suggesting that tumor suppressors or oncogenes could act as a double-edged sword in cancer progression under some specific conditions. For instance, microRNA-149, a p53 upregulated microRNA, was reported to act as an oncogenic regulator in human melanoma by targeting glycogen synthase kinase-3α [10].
In conclusion, our work demonstrates the existence of fusion circBA1 and its tumor-suppressive role in BCR-ABL positive CML cells, provides new insights into the CML progression, and expands the current knowledge regarding fusion genes produced from chromatin translocations.
DECLARATIONS
AUTHORSHIP
JKZ and JLY designed and supervised the study and experiments; JW, JKZ, HLM, and WRL performed the experiments. JW, JKZ, and JLY analyzed the data; JW, JKZ, and YP wrote the manuscript; all authors have read and approved the final manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the Ethics Committee of West China Hospital. All human samples were obtained with informed consent.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
FUNDING
This work is supported by grants from the National Natural Science Foundation of China (81900156), the China Postdoctoral Science Foundation (2018M640927), and the Post-Doctor Research Project, West China Hospital, Sichuan University (2018HXBH022).
ACKNOWLEDGEMENTS
We would like to thank Prof. Jeremy E. Wiluse for providing us the plasmid pcDNA3.1(+) Laccase2 MCS Exon.
Open Research
DATA AVAILABILITY STATEMENT
Data can be requested from the corresponding author upon reasonable request.




