LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer
Abstract
Altered metabolism is a hallmark of cancer, and the reprogramming of energy metabolism has historically been considered a general phenomenon of tumors. It is well recognized that long noncoding RNAs (lncRNAs) regulate energy metabolism in cancer. However, lncRNA-mediated posttranslational modifications and metabolic reprogramming are unclear at present. In this review, we summarized the current understanding of the interactions between the alterations in cancer-associated energy metabolism and the lncRNA-mediated posttranslational modifications of metabolic enzymes, transcription factors, and other proteins involved in metabolic pathways. In addition, we discuss the mechanisms through which these interactions contribute to tumor initiation and progression, and the key roles and clinical significance of functional lncRNAs. We believe that an in-depth understanding of lncRNA-mediated cancer metabolic reprogramming can help to identify cellular vulnerabilities that can be exploited for cancer diagnosis and therapy.
Abbreviations
-
- 1,3-BPG
-
- 1,3-bisphosphoglycerate
-
- 3-PG
-
- 3-phosphoglycerate
-
- AGPG
-
- actin gamma 1 pseudogene
-
- AML
-
- acute myeloid leukaemia
-
- AMPK
-
- adenosine monophosphate-activated protein kinase
-
- ANRIL
-
- antisense noncoding RNA in the INK4 locus
-
- ATG
-
- autophagy-related gene 5
-
- ATP
-
- adenosine-triphosphate
-
- BRK
-
- breast tumor kinase
-
- CamK-A
-
- calcium-dependent kinase activation
-
- CASC9
-
- cancer susceptibility candidate 9
-
- CDK6
-
- cyclin-dependent kinase 6
-
- ceRNA
-
- competing endogenous RNA
-
- circRNA
-
- circular RNA
-
- CRC
-
- colorectal cancer
-
- DHX9
-
- DEAH (Asp-Glu-Ala- His) box helicase proteins 9
-
- DRAIC
-
- downregulated RNA in cancer
-
- ENO1
-
- enolase 1
-
- F-2,6-BP
-
- fructose-2,6-bisphosphate
-
- F6P
-
- fructose 6-phosphate
-
- FEZF1-AS1
-
- Fez family zinc finger protein 1 antisense ribonucleic acid 1
-
- FGFR1
-
- fibroblast growth factor receptor type 1
-
- FUS
-
- fused in sarcoma
-
- G3BP1
-
- Ras-GTPase-activating protein-binding protein 1
-
- GBCDRlnc1
-
- gallbladder cancer drug resistance-associated lncRNA1
-
- GDH1
-
- glutamate dehydrogenase 1
-
- GLCC1
-
- glycolysis-associated lncRNA of colorectal cancer 1
-
- GLS-AS
-
- antisense lncRNA of glutaminase
-
- GLUT1
-
- glucose transporter isoform 1
-
- HCC
-
- hepatocellular carcinoma
-
- HIF-1α
-
- hypoxia-inducible factor-1α
-
- HISLA
-
- HIF-1α-stabilizing lncRNA
-
- HK2
-
- hexokinase 2
-
- HNF4α
-
- hepatocyte nuclear factor 4 alpha
-
- HSP90
-
- heat shock protein 90
-
- HULC
-
- highly up-regulated in liver cancer
-
- IGF2BP2
-
- insulin-like growth factor 2 mRNA-binding protein 2
-
- IKK
-
- IκB kinase
-
- IκBα
-
- inhibitor kappa B alpha
-
- KRT19P3
-
- keratin 19 pseudogene 3
-
- LDHA
-
- lactate dehydrogenase A
-
- lincRNA
-
- long intergenic noncoding RNA
-
- LINK-A
-
- long intergenic noncoding RNA for kinase activation
-
- LINP1
-
- lncRNA in nonhomologous end joining pathway 1
-
- LINRIS
-
- long intergenic noncoding RNA for IGF2BP2 stability
-
- LKB1
-
- liver kinase B1
-
- lncRNA
-
- long noncoding RNA
-
- LRRK2
-
- leucine-rich repeat kinase 2
-
- m6A
-
- N6-methyladenosine
-
- MACC1
-
- metastasis-associated in colon cancer-1
-
- MALAT1
-
- metastasis-associated lung adenocarcinoma transcript 1
-
- MDM2
-
- murine double minute 2
-
- MEG3
-
- maternally expressed gene 3
-
- miRNA
-
- microRNA
-
- mTOR
-
- mammalian target of rapamycin
-
- NADH
-
- nicotinamide adenine dinucleotide
-
- NADPH
-
- nicotinamide adenine dinucleotide phosphate
-
- NBR2
-
- neighbour of BRCA1 gene 2
-
- NF-κB
-
- nuclear factor κB
-
- NSCLC
-
- non-small cell lung cancer
-
- OXPHOS
-
- oxidative phosphorylation
-
- PFK1
-
- phosphofructokinase-1
-
- PFKFB3
-
- 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3
-
- PGK1
-
- phosphoglycerate kinase 1
-
- PHD2
-
- proline hydroxylase domain 2
-
- PI3K
-
- phosphoinositide-3-kinase
-
- PKM2
-
- pyruvate kinase muscle isozyme M2
-
- PNCK
-
- pregnancy upregulated non-ubiquitous calmodulin kinase
-
- PRAL
-
- p53 regulation-associated lncRNA
-
- SIRT1
-
- Sirtuin 1
-
- TCA
-
- tricarboxylic acid
-
- VHL
-
- von Hippel-Lindau
-
- α-KG
-
- alpha-ketoglutarate
1 BACKGROUND
Altered metabolism is a hallmark of cancer, and the reprogramming of energy metabolism has historically been considered a general phenomenon of tumors [1-3]. Specifically, tumor cells usually undergo energy metabolic reprogramming to satisfy the requirements for proliferation and survival in unfavourable environments, ultimately leading to cancer progression [4]. In addition to energy, tumor cells also require building blocks, such as the cofactors nicotinamide adenine dinucleotide phosphate (NADPH) and nicotinamide adenine dinucleotide (NADH), to support the housekeeping processes, growth, and proliferation in a dynamic microenvironment [5, 6]. Malignant tumor cells exhibit a highly regulated programme of metabolic plasticity [7]. Metabolic reprogramming and malignant transformation are complex and multistep processes that are associated with the accumulation of numerous molecular alterations [8]. On the one hand, altered metabolic signalling networks are driven by aberrant genetic alterations, including mutations, loss of heterozygosity, deletions, insertions, aneuploidy, etc., that lead to the activation of oncogenes and inactivation of tumor suppressors [9, 10]. On the other hand, epigenetic mechanisms, including DNA and histone methylation, noncoding RNA regulation, posttranslational modifications, etc., are also critical mediators of the functional changes that drive and maintain the malignant phenotypes and metabolic reprogramming [11-13].
Long noncoding RNAs (lncRNAs) are a heterogeneous group of non-protein-coding transcripts with the lengths of greater than 200 nucleotides [14-16]. LncRNAs are emerging regulators that are involved in the gene expression, diverse physiological and pathological processes [12, 17, 18]. Increasing evidence shows that lncRNAs play complex and precise regulatory roles in cancer initiation and progression by acting as oncogenes or tumor suppressors [19, 20]. They can regulate not only the proliferation, differentiation, invasion, and metastasis but also metabolic reprogramming of cancer cells [12, 21, 22]. Moreover, accumulating evidence suggests that lncRNAs play important functional roles in modulating the transcription and translation of metabolism-related genes, serving as decoys, scaffolds, competing endogenous RNAs (ceRNAs), etc., ultimately leading to cancer metabolic reprogramming [23, 24]. In addition, recent studies by our group and other groups also suggested that lncRNAs promote energy metabolism and cancer progression through posttranslational modifications of key metabolism-related proteins, including the ubiquitination, phosphorylation, and acetylation [25-29].
In this review, we discuss the significant roles of lncRNAs in the reprogramming of energy metabolism and the progression of cancers. In particular, we focus on lncRNA-mediated posttranslational modifications of metabolic enzymes, metabolism-related transcription factors, and other proteins in relevant signalling pathways, highlighting recent important insights into the mechanisms regulating cancer metabolism.
2 LncRNA-MEDIATED POSTTRANSLATIONAL MODIFICATION OF METABOLIC ENZYMES
Tumor cell metabolism is reprogrammed from oxidative phosphorylation (OXPHOS) to aerobic glycolysis even when oxygen is sufficient, a phenomenon called the “Warburg effect” [30]. Therefore, the abnormal expression and regulation of related enzymes in the glycolytic pathway are vital for tumor development [31]. As shown in Figure 1 and Supplementary Table S1, we first summarize the available studies on lncRNA-mediated posttranslational modifications of several metabolic enzymes in aerobic glycolysis. The relationship between lncRNAs and metabolic enzymes is complicated and flexible, because they can influence each other. The specific mechanisms of their actions and relationship deserve in-depth studies in the future.
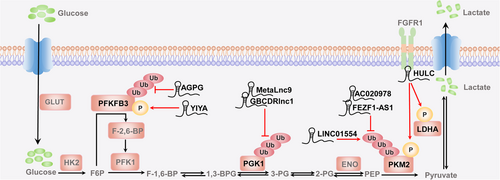
2.1 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 (PFKFB3)
In aerobic glycolysis, PFKFB3 converts fructose 6-phosphate (F6P) into fructose-2,6-bisphosphate (F-2,6-BP), which can act as an allosteric activator of the rate-limiting enzyme phosphofructokinase-1 (PFK1) [25]. Studies have identified that lncRNA-mediated reprogramming of energy metabolism is closely related to the PFKFB3 ubiquitination and phosphorylation in cancer [25, 32]. Moreover, our recent study revealed that the lncRNA actin gamma 1 pseudogene (AGPG) specifically bound to the C-terminal domain of PFKFB3, which stimulated the stable expression of PFKFB3 and activated the glycolytic flux by inhibiting the ubiquitination of PFKFB3 at Lys302 and its subsequent proteasome-dependent degradation, leading to metabolic reprogramming in esophageal squamous cell carcinoma cells [25]. In addition, PFKFB3 could also be phosphorylated by the lncRNA YIYA (LINC00538), which was associated with cyclin-dependent kinase 6 (CDK6) and facilitated its binding to cyclin D3. Thus, YIYA regulated the CDK6-dependent phosphorylation of PFKFB3 in a cell cycle-independent manner, which led to the enhanced conversion of F6P to F-2,6-BP and promoted the reprogramming of glucose metabolism and the growth of breast cancer [32].
2.2 Phosphoglycerate kinase 1 (PGK1)
PGK1 is the first crucial metabolic enzyme mediating adenosine-triphosphate (ATP) production via glycolysis and catalysing the conversion of 1,3-bisphosphoglycerate (1,3-BPG) to 3-phosphoglycerate (3-PG) [33]. The activity of PGK1 is mediated by different posttranslational modifications. For example, PGK1 can frequently be phosphorylated, ubiquitinated, and acetylated at multiple sites to regulate tumor metabolism and cancer progression [34-37]. A recent study showed that MetaLnc9 (LINC00963) played a dominant role in the posttranslational activity of PGK1 and stabilized PGK1 by blocking its ubiquitin-mediated degradation, allowing it to exert its carcinogenic activity and accelerate the metastasis of non-small cell lung cancer (NSCLC) cells via the phosphoinositide-3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) signalling [36]. Similarly, gallbladder cancer drug resistance-associated lncRNA1 (GBCDRlnc1), which served as a “decoy molecule”, posttranslationally regulated the level of PGK1 by directly preventing its ubiquitin-mediated degradation in gallbladder cancer. Expression of the autophagy initiator autophagy-related gene 5 (ATG5)-ATG12 conjugate was ultimately up-regulated through the formation of the GBCDRlnc1/PGK1/ATG5-ATG12 complex, which induced autophagy and drug resistance in gallbladder cancer cells [37].
2.3 Pyruvate kinase muscle isozyme M2 (PKM2)
PKM2 catalyses the conversion of phosphoenolpyruvate to pyruvate and generates ATP in the rate-limiting step in glycolysis, which switches glucose metabolism from the normal respiratory chain to lactate production in tumor cells [38, 39]. Coincidentally, PKM2 activity is also regulated by posttranslational modifications, including O-GlcNAcylation, acetylation, phosphorylation, ubiquitination, etc [40-45]. The lncRNAs AC020978, Fez family zinc finger protein 1 antisense ribonucleic acid 1 (FEZF1-AS1), and LINC01554 can reprogram tumor metabolism by regulating the stability of PKM2 via the ubiquitin-mediated proteasome pathway. In detail, AC020978 specifically modulated the stability of the PKM2/hypoxia-inducible factor-1α (HIF-1α) complex, furthered the action of HIF-1α on its target genes, thus forming a positive feedback loop related to tumor-specific metabolism and endowing NSCLC cells with an aggressive phenotype [40]. Additionally, FEZF1-AS1 bound to and increased the stability of PKM2, increasing cytoplasmic and nuclear PKM2 levels. Thus, FEZF1-AS1 promoted the aerobic glycolysis and accelerated the proliferation and metastasis of colorectal cancer (CRC) cells [41]. In contrast, the lncRNA LINC01554 accelerated ubiquitin-mediated degradation of PKM2 to inhibit aerobic glycolysis in hepatocellular carcinoma (HCC) cells via the AKT/mTOR signalling pathway [42]. Moreover, the lncRNA highly up-regulated in liver cancer (HULC) functioned as an adaptor molecule and enhanced the interactions of lactate dehydrogenase A (LDHA) and PKM2 with the intracellular domain of the upstream kinase fibroblast growth factor receptor type 1 (FGFR1), leading to the elevated phosphorylation of LDHA and PKM2, which consequently enhanced glycolysis and increased the proliferation of liver cancer cells [46].
3 LncRNA-MEDIATED POSTTRANSLATIONAL MODIFICATION OF TRANSCRIPTION FACTORS
In addition to directly regulating energy metabolism via posttranslational modification of metabolic enzymes, lncRNAs can also regulate metabolism-related transcription factors to reprogram energy metabolism since the regulation of transcription factors upon their targets is influenced by lncRNAs [47]. LncRNAs regulate the activity of these proteins through hydroxylation, phosphorylation, and ubiquitination, thus enhancing the expression of metabolism-related enzymes and promoting the malignant progression of tumors (Figure 2 and Supplementary Table S1). Therefore, we next discuss the recently reported lncRNAs and the mechanisms by which they regulate representative factors, including HIF-1α, c-Myc, and nuclear factor κB (NF-κB).
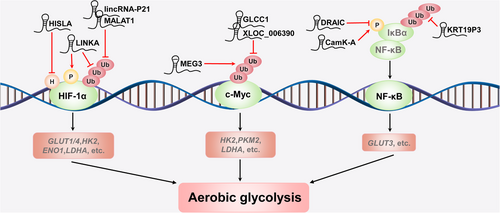
3.1 HIF-1α
HIF-1α plays a key role in the reprogramming of cancer metabolism by activating the transcription of genes that encode glucose transporters and glycolytic enzymes [48, 49]. Accumulating evidence suggests that lncRNAs play important functional roles in regulating HIF-1α activity and stability. For example, Su et al. [50] found that the lncRNA cancer susceptibility candidate 9 (CASC9) reprogramed glycolytic metabolism and stimulated the tumorigenic behaviour of nasopharyngeal cancer cells by interacting with HIF-1α and increasing the abundance of HIF-1α. Chen et al. [51] found that HIF-1α-stabilizing lncRNA (HISLA) inhibited the hydroxylation and degradation of HIF-1α and promoted the aerobic glycolysis in breast cancer cells by blocking the interaction between proline hydroxylase domain 2 (PHD2) and HIF-1α. More interestingly, by activating breast tumor kinase (BRK) phosphorylation at Tyr351, long intergenic noncoding RNA for kinase activation (LINK-A) mediated HIF-1α phosphorylation at Tyr565, which prevented the degradation of HIF-1α under normoxic conditions; moreover, leucine-rich repeat kinase 2 (LRRK2) recruited by LINK-A could also phosphorylate HIF-1α at Ser797, which led to the activation of HIF-1α target genes. The above series of LINK-A-mediated posttranslational modifications promoted glycolytic reprogramming and tumorigenesis in triple-negative breast cancer [52].
Under normoxic conditions, HIF-1α is hydroxylated by PHD at Pro402 and/or Pro564 [53]. After hydroxylation, HIF-1α binds to von Hippel-Lindau (VHL) and undergoes VHL-mediated ubiquitination and degradation via the proteasome pathway. However, the activity of PHD is suppressed under hypoxic conditions. A recent study reported that accumulation of HIF-1α resulted in high expression of PHD, which rapidly induced transcription of long intergenic noncoding RNA (lincRNA)-p21 [54]. Interestingly, highly expressed lincRNA-p21 removed the polyubiquitin chain at Hyp564 installed by VHL and decreased HIF-1α degradation via competitively binding with HIF-1α. As discussed above, the formation of a positive HIF-1α-lincRNA-p21-HIF-1α feedback loop ensured that HIF-1α was highly induced in hypoxia, activating glucose transporter isoform 1 (GLUT1) and LDHA and enhancing the anaerobic glycolysis pathway, which promoted tumor growth [54]. Similarly, the lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) increased the expression of core glycolytic enzymes, including hexokinase 2 (HK2), enolase 1 (ENO1), and GLUT4, by reducing the VHL-mediated ubiquitination and subsequent degradation of HIF-1α [20, 55].
3.2 c-Myc
The c-Myc protein acts as a “master regulator” of cellular metabolism and cancer progression by stimulating many metabolic changes that result in malignant transformation [56]. Additionally, lncRNAs have been reported to be involved in the regulation of cancer energy metabolism via the posttranslational modification of c-Myc. Several lncRNAs are significantly overexpressed and function as oncogenes in CRC or pancreatic cancer, including glycolysis-associated lncRNA of colorectal cancer 1 (GLCC1), long intergenic noncoding RNA for insulin-like growth factor 2 mRNA-binding protein 2 stability (LINRIS), and lncRNA XLOC_006390. Mechanistically, the lncRNA GLCC1 protected c-Myc from ubiquitination by directly interacting with the chaperone heat shock protein 90 (HSP90) and further specified the transcriptional modification pattern of c-Myc target genes such as LDHA, consequently reprogramming glycolytic metabolism for CRC cell proliferation [57]. The lncRNA LINRIS bound to insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2), repressed its ubiquitination and prevented its degradation via the autophagy-lysosome pathway. The LINRIS-IGF2BP2 axis promoted the translation of its target gene c-Myc and accelerated the malignant proliferation of CRC cells by up-regulating aerobic glycolysis [26]. The lncRNA XLOC_006390 stablized c-Myc by preventing its ubiquitination, increasing the expression of glutamate dehydrogenase 1 (GDH1), and subsequently stimulating the production of alpha-ketoglutarate (α-KG). Excess α-KG supplied the tricarboxylic acid (TCA) cycle and facilitated glutamate metabolism, promoting pancreatic cancer growth [58].
In contrast, the lncRNA maternally expressed gene 3 (MEG3) and the lncRNA antisense lncRNA of glutaminase (GLS-AS) were down-regulated and functioned as tumor suppressors in cancer. Mechanistically, MEG3 overexpression enhanced ubiquitin-dependent degradation of c-Myc and repressed c-Myc target genes involved in the glycolytic pathway, including LDHA, PKM2, and HK2 [59]. Similarly, the lncRNA GLS-AS restricts glutaminase expression at the posttranscriptional level by binding with the glutaminase pre-mRNA, resulting in the formation of a double-stranded RNA. Furthermore, GLS-AS reduce Myc expression by disrupting the glutaminase-mediated stabilization of Myc. Conversely, in pancreatic cancer, Myc up-regulated the expression of GLS-AS under nutritional stress, thus forming a mutual feedback loop to inhibit metabolic reprogramming [60].
3.3 NF-κB
NF-κB is an inducible transcription factor that plays a core role in diverse biological processes, including cellular metabolism, epithelial-to-mesenchymal transition, invasion, angiogenesis, metastasis, therapeutic resistance, and other processes [61]. Aberrant NF-κB signalling has been verified to be involved in the pathogenesis of most human cancers [62]. Direct regulation of cell metabolism via the NF-κB pathway has been addressed by several recent studies [63, 64]. In particular, our recent study validated that lncRNA for calcium-dependent kinase activation (CamK-A) regulated Ca2+ signalling-mediated remodelling of tumor microenvironment and reprogramming of energy metabolism through posttranslational modification and NF-κB activation [27]. Mechanistically, CamK-A bound both the calmodulin-dependent kinase pregnancy up-regulated non-ubiquitous calmodulin kinase (PNCK) and the C-terminal SH3-like region of inhibitor kappa B alpha (IκBα). Thus, CamK-A promotes PNCK-induce phosphorylation of IκBα at Ser32 under hypoxic conditions, which further induce IκBα degradation and NF-κB activation. In contrast, several NF-κB-related lncRNAs are down-regulated and function as tumor suppressors during cancer progression. For example, Saha et al. [65] reported that the downregulated RNA in cancer (DRAIC) interacted with the subunits of the IκB kinase (IKK) complex and subsequently inhibited prostate cancer progression by repressing IκBα phosphorylation and NF-κB activation. Additionally, the lncRNA keratin 19 pseudogene 3 (KRT19P3) inhibited gastric cancer cell growth and metastasis by modulating IκBα deubiquitination and inactivating NF-κB signalling [66].
4 LncRNA-MEDIATED POSTTRANSLATIONAL MODIFICATION OF OTHER PROTEINS INVOLVED IN METABOLIC PATHWAYS
Alterations in proteins involved in metabolism are accompanied by alterations in metabolic pathways, which will eventually become carcinogenic signalling pathways or be activated by carcinogenic factors [8]. Through these abnormal metabolic pathways, tumor cells can obtain the necessary materials and energy required for continuous proliferation [67]. In addition to directly regulating the posttranslational modification of key metabolic enzymes, lncRNAs can also indirectly modulate metabolic pathways through posttranslational modifications (Figure 3 and Supplementary Table S1). Among the observed effects, functional alterations in the adenosine monophosphate-activated protein kinase (AMPK) pathway, PI3K/AKT pathway and p53 pathway are particularly prominent.
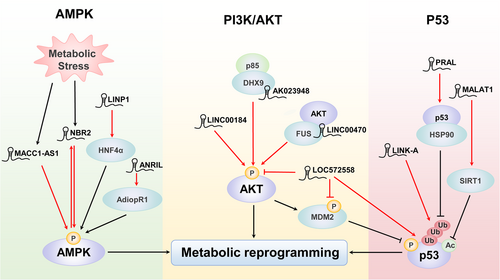
4.1 AMPK signalling pathway
AMPK is a critical cellular energy sensor that mainly functions as a metabolic checkpoint to restore energy balance in response to different types of metabolic stress [68]. AMPK is activated by phosphorylation, which maintains energy balance, redox homeostasis and cell survival by regulating glycolysis, fatty acid metabolism, antioxidant reactions, and other processes [69, 70]. Recent studies have showed that AMPK phosphorylation is epigenetically regulated by different signalling pathways involving different lncRNAs, including metastasis-associated in colon cancer-1 (MACC1)-AS1, antisense noncoding RNA in the INK4 locus (ANRIL), lncRNA in nonhomologous end joining pathway 1 (LINP1) and neighbour of BRCA1 gene 2 (NBR2). MACC1-AS1 is the antisense lncRNA transcript of the transcriptional regulator MACC1, which is recognized as an oncogene in cancer. MACC1-AS1 mediated metabolic plasticity by up-regulating MACC1 and subsequently enhanced glycolysis and antioxidative capabilities via the phosphorylation of AMPK and the translocation of Lin28 (an evolutionarily conserved RNA-binding protein), ultimately accelerating the growth of gastric cancer [71]. In addition, the lncRNA ANRIL promoted the survival of malignant cells and cellular glucose metabolism, accelerating acute myeloid leukaemia (AML) progression through AMPK phosphorylation and forming the AdipoR1/AMPK/Sirtuin 1 (SIRT1) pathway [72]. In addition, the lncRNA LINP1 regulated AML progression via increasing AMPK phosphorylation mediated through the hepatocyte nuclear factor 4 alpha (HNF4α)/AMPK/WNT5A signalling pathway [73]. In contrast, Liu et al. [74] proposed a feed-forward model of NBR2-AMPK regulation, in which the lncRNA NBR2 was induced by the liver kinase B1 (LKB1)-AMPK pathway under energy stress, and in turn NBR2 interacted with AMPK and promoted its phosphorylation.
4.2 PI3K/AKT signalling pathway
PI3K/AKT plays a key role in diverse cellular processes in cancer, including cell survival, proliferation, angiogenesis, and energy metabolism [75, 76]. Evidence has revealed that numerous positive regulators, including growth factors, regulatory proteins, and microRNAs (miRNAs), promote the overactivation of AKT signalling [77]. Some lncRNAs, such as LOC572558, LINC00184, LINC00470 (also known as C18orf2) and AK023948, have been associated with AKT phosphorylation and energy metabolic reprogramming in diseases, especially in cancer. One study showed that LINC00184 modulated glycolysis and mitochondrial oxidative phosphorylation in oesophageal cancer cells through inducing AKT phosphorylation [78]. In addition, Liu et al. [79] demonstrated that LINC00470 was a positive regulator of AKT activation in glioblastoma cells. Mechanistically, LINC00470 bound to the RNA-binding protein fused in sarcoma (FUS) and AKT, forming a ternary complex that anchors FUS in the cytoplasm and increases AKT phosphorylation. AKT activation by LINC00470 inhibited HK1 ubiquitination and promoted aerobic glycolysis and glioblastoma tumorigenesis. AK023948 is also reported to function as a positive regulator of AKT. AK023948 was required for the interaction between DEAH (Asp-Glu-Ala- His) box helicase proteins 9 (DHX9) and p85 to enhance p85 stability and promote AKT activity, which may contribute to breast tumor progression [80]. Overexpression of LOC572558, however, resulted in the dephosphorylation of AKT and murine double minute 2 (MDM2) while the phosphorylation of p53, which inhibited cell proliferation and tumor growth in bladder cancer through the AKT-MDM2-p53 axis [81].
4.3 p53 signalling pathway
In addition to the functions of the above basic metabolic pathways, the functions of p53-related signalling pathways in metabolism cannot be ignored [82, 83]. Kruiswijk et al. [84] proposed that cell metabolism was a noncanonical p53-controlled process, and this idea had attracted considerable attention. Importantly, activation of p53 reprograms tumor glucose metabolism by reversing the Warburg effect, which negatively affected the carcinogenic metabolism of cancer cells and prevented the development of aggressive tumor phenotypes [84, 85]. Recently, the importance of lncRNAs and p53 in cancer metabolic reprogramming has been confirmed [29, 86]. Mao et al. [87] confirmed that the interaction between the lncRNA P53RRA and Ras-GTPase-activating protein-binding protein 1 (G3BP1) prevented p53 from forming the G3BP1 complex, which repressed the transcription of several metabolism-related genes. In addition, various lncRNAs, such as LOC572558, p53 regulation-associated lncRNA (PRAL), LINK-A and MALAT1, also regulated p53 through the posttranslational modifications. In terms of ubiquitination, PRAL enhanced the stability of p53 by competitively binding with it and blocking its MDM2-dependent degradation. The three stem-loop motifs at the 5' end of PRAL promoted the interaction between p53 and HSP90 [88]. However, LINK-A enhanced the K48-polyubiquitination-mediated degradation of p53 [89]. In addition, Chen et al. [90] suggested that MALAT1 suppressed cell apoptosis and promoted cell proliferation by enhancing the SIRT1-mediated deacetylation of p53 and then down-regulating its downstream target genes. Together, the above findings indicate that lncRNA-mediated posttranslational modifications are crucial to the p53-related metabolic pathways.
5 CONCLUSIONS
The discovery and functional studies of metabolism-associated lncRNAs expand our understanding of the regulatory mechanisms in cancer. Importantly, the functions of these identified lncRNAs are critical to tumorigenesis, tumor progression, and metabolic reprogramming. As technological advances have flourished in the past decade, understanding the precise contributions of lncRNA-mediated metabolic reprogramming has become easier. In this review, we highlight the flexible and complex energy metabolism network that is controlled by lncRNA-mediated posttranslational modifications, one of the most rapid manners by which cells respond to external and internal stimuli, and is a hub of many cellular signalling pathways. Posttranslational modifications alter proteins by adding or removing different functional groups, thereby modulating the protein structure and function. In addition, the suite of epigenetic modifications mediated by lncRNAs is complex and diverse, and includes ubiquitination, phosphorylation and acetylation, all of which can lead to the reprogramming of energy metabolism. On the contrary, RNA epigenetic modification and its biological significance, such as N6-methyladenosine (m6A), are emerging research frontiers in tumor biology and deserve extensive study. In general, the current research on this topic in cancer is limited, and the existing knowledge has not been fully translated into clinical applications. However, the current findings offer new scientific ideas and perspectives on cancer energy metabolism.
Recent studies of cancer metabolism have focused on assessing metabolic phenotypes in the tumor microenvironment, especially the tumor immune microenvironment. These studies have expanded the scope of metabolic dependencies beyond the classical pathways that dominate metabolism in cultured cells. Accumulating evidence from recent studies indicates that alterations in the tumor microenvironment can drive tumorigenesis. In addition, studies have indicated that cell metabolism influences T cell antitumor activity, and strategies to modulate metabolism can improve the tumor immune response. We believe that lncRNA-mediated metabolic reprogramming also plays critical roles in remodelling of the tumor microenvironment and in tumor immune escape. In addition to lncRNA, the role of circular RNAs (circRNAs) in tumor metabolic remodelling and immune microenvironment has also attracted more attention. Thus, identification of these functional ncRNAs, and investigation of their biological roles and underlying regulation mechanisms in tumor microenvironment remodelling need to be further studied.
Taken together, in-depth clarification of the common lncRNA-mediated regulatory mechanisms in metabolic reprogramming will provide a deeper understanding of tumor development and cellular vulnerabilities. Moreover, the involved lncRNAs can also be exploited as reliable diagnostic biomarkers and therapeutic targets in cancer.
ACKNOWLEDGEMENTS
Not applicable.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
COMPETING INTERESTS
The authors declare that they have no competing interests.
FUNDING
This study was supported by the National Key R&D Program of China (2018YFC1313304 and 2018YFC1313300) and the National Natural Science Foundation of China (82073112, 82022052 and 81871951).
AUTHORS' CONTRIBUTIONS
Conceptualization: HQJ and RHX. Article writing and editing: YTT, JFL, TL, JJL and HQJ. Supervision: HQJand RHX. All authors read and approved the final manuscript.




