Single-cell RNA sequencing in breast cancer: Understanding tumor heterogeneity and paving roads to individualized therapy
Abstract
Single-cell RNA sequencing (scRNA-seq) is a novel technology that allows transcriptomic analyses of individual cells. During the past decade, scRNA-seq sensitivity, accuracy, and efficiency have improved due to innovations including more sensitive, automated, and cost-effective single-cell isolation methods with higher throughput as well as ongoing technological development of scRNA-seq protocols. Among the variety of current approaches with distinct features, researchers can choose the most suitable method to carry out their research. By profiling single cells in a complex population mix, scRNA-seq presents great advantages over traditional sequencing methods in dissecting heterogeneity in cell populations hidden in bulk analysis and exploring rare cell types associated with tumorigenesis and metastasis. scRNA-seq studies in recent years in the field of breast cancer research have clustered breast cancer cell populations with different molecular subtypes to identify distinct populations that may correlate with poor prognosis and drug resistance. The technology has also been used to explain tumor microenvironment heterogeneity by identifying distinct immune cell subsets that may be associated with immunosurveillance and are potential immunotherapy targets. Moreover, scRNA-seq has diverse applications in breast cancer research besides exploring heterogeneity, including the analysis of cell-cell communications, regulatory single-cell states, immune cell distributions, and more. scRNA-seq is also a promising tool that can facilitate individualized therapy due to its ability to define cell subsets with potential treatment targets. Although scRNA-seq studies of therapeutic selection in breast cancer are currently limited, the application of this technology in this field is prospective. Joint efforts and original ideas are needed to better implement scRNA-seq technologies in breast cancer research to pave the way for individualized treatment management. This review provides a brief introduction on the currently available scRNA-seq approaches along with their corresponding strengths and weaknesses and may act as a reference for the selection of suitable methods for research. We also discuss the current applications of scRNA-seq in breast cancer research for tumor heterogeneity analysis, individualized therapy, and the other research directions mentioned above by reviewing corresponding published studies. Finally, we discuss the limitations of current scRNA-seq technologies and technical problems that remain to be overcome.
Abbreviations
-
- CDK4/6
-
- Cyclin-dependent kinases 4 and 6
-
- CI
-
- Confidential interval
-
- CSC
-
- Cancer stem cells
-
- CTC
-
- Circulating tumor cells
-
- EMT
-
- Epithelial–mesenchymal transition
-
- ER
-
- Estrogen receptor
-
- FACS
-
- Fluorescence-activated cell sorting
-
- HER2
-
- Human epidermal growth factor receptor2
-
- HR
-
- Hazard ratio
-
- ICOSL
-
- Inducible T-cell co-stimulator ligand
-
- IMC
-
- Immunosuppressive immature myeloid cell
-
- LCM
-
- Laser capture microdissection
-
- LP
-
- Luminal progenitor
-
- OXPHOS
-
- Mitochondrial oxidative phosphorylation
-
- pCR
-
- Pathologic complete response
-
- PCR
-
- Polymerase chain reaction
-
- PD-L1
-
- Programmed death-ligand 1
-
- PR
-
- Progesterone receptor
-
- RNA-seq
-
- RNA sequencing
-
- scRNA-seq
-
- Single-cell RNA-sequencing
-
- TMB
-
- Tumor mutation burden
-
- TNBC
-
- Triple-negative breast cancer
-
- TRM
-
- Pathologic complete response
-
- TRM
-
- Tissue-resident memory T
-
- UMIs
-
- Unique molecular identifiers
1 BACKGROUND
Traditional RNA sequencing (RNA-seq) is performed using bulk RNA extracted from homogenized tissues or large cell populations to ensure sufficient RNA for subsequent analyses. However, bulk RNA-seq only provides an average number of gene expression in the pooled population of diverse cells and cannot capture the widespread transcriptome heterogeneity in the cell population [1]. Therefore, analyses of bulk gene expression data cannot identify distinct cell types that express certain genes but instead provide a virtual average of the multiple cellular components, which may represent very little information about the specific cell type present [1, 2].
Due to technology advances, gene expression analysis can now be performed at a much higher resolution [3, 4]. The expression level of every gene, even in a single cell, can now be defined. In contrast to bulk RNA analysis, the recently developed single-cell RNA-sequencing (scRNA-seq) provides high-throughput, and high-resolution transcriptomic analyses of individual cells. By isolating single cells, capturing their transcripts, and generating sequencing libraries at the single-cell level, scRNA-seq can reveal the state and function of single cells. scRNA-seq was first introduced by Tang et al. [5] in 2009. The first scRNA-seq studies were conducted in a number of 10 ∼ 100 cells [6-8]. With the evolution of technology, these methods have been gradually refined and new approaches have been developed and the transcriptomic analyses of up to tens of thousands of individual cells for a single project have been achieved [9, 10].
Breast tumors contain a heterogeneous mix of cells that includes cancer, vascular, immune, and fibroblast cell types [11]. ScRNA-seq allows a deeper understanding of the diversity of cell states and the heterogeneity of cell populations, making it a useful tool for dissecting the properties of the multiple cell types within and surrounding breast tumors. The application of scRNA-seq can also improve our understanding on the mechanisms of oncogenesis and metastasis in breast cancer to pave the way for individualized therapy. Due to the prominent above-mentioned advantages, scRNA-seq has become a booming technology in breast cancer studies in recent years. Researchers have utilized scRNA-seq to analyze tumor heterogeneity in breast cancer of different molecular subtypes and have identified cell clusters related to poor prognosis or therapeutic response [12-14]. Single-cell profiling of diverse immune cells in the tumor microenvironment of breast cancer has revealed specific immune cell subpopulations that may be potential immunotherapy targets [15-17]. Studies focused on cell-cell communications, regulatory single-cell states, and immune cell distributions in breast cancer have also been conducted by scRNA-seq [18-21]. Furthermore, researchers have used scRNA-seq to analyze the association between therapeutic response and specific infiltrated immune cells in the tumor environment [22, 23]. However, there is still a research gap to be filled by scRNA-seq. For instance, intratumoral heterogeneity that hampers the efficacy of targeted therapy needs to be thoroughly studied, rationale strategies are needed to identify drug-resistant cell populations associated with poor prognosis to achieve long-term treatment efficacy and combinatorial therapeutic strategies are required to co-target the multiple activated pathways. Although scRNA-seq showed an incomparable advantage over bulk RNA-seq in cancer research, there is still room for improvement. Many technical problems need to be tackled to enable scRNA-seq to provide a better service for cancer research, such as protection of cell viability during single-cell isolation, a better combination of spatial information and sequencing data, and more.
Here, we reviewed the current scRNA-seq methods and discussed their corresponding advantages and disadvantages to provide researchers with a reference for the selection of suitable approaches. We then discussed the current applications of scRNA-seq in breast cancer research and analyzed its application prospects and technical barriers that need to be overcome in the future.
2 TECHNICAL APPROACHES TO SCRNA SEQUENCING
2.1 scRNA-seq workflow
The workflow of the critical steps in a typical scRNA-seq experiment is shown in Figure 1. Compared to traditional bulk RNA analyses, single cells must be isolated from the tumor tissues at the beginning of scRNA-seq. Second, specific protocols are needed to perform mRNA reverse transcription and cDNA amplification with high efficiency and low biases as the quantity of RNA in a single cell is lower than that in bulk RNA analyses. New strategies have also been developed to better analyze data from scRNA-seq because previously developed tools for bulk RNA-seq were not appropriate for these complex data analysis. A comparison of scRNA-seq and bulk RNA analysis workflows is shown in Figure 2.
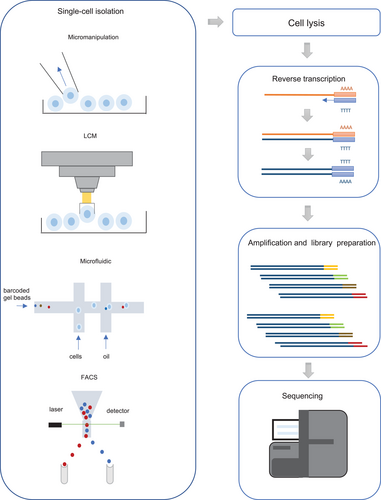
Illustration of single-cell RNA-sequencing (scRNA-seq) experiments.
A typical scRNA-seq workflow includes most of the following steps: 1) single-cell isolation by micromanipulation, laser-capture microdissection (LCM), microfluidics, fluorescence-activated cell sorting (FACS) 2) cell lysis 3) reverse transcription of mRNA to cDNA 4) cDNA amplification and library preparation 5) sequencing
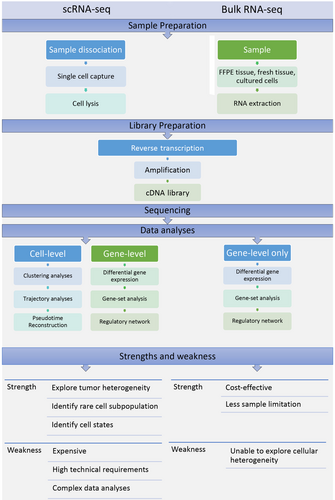
Workflow of a typical single-cell RNA-sequencing experiment and traditional bulk RNA analyses.
The main difference between the workflows of single-cell RNA-sequencing and traditional bulk RNA analyses lies in the first step. In single-cell RNA-sequencing, a single cell must be isolated from the samples at the beginning while in traditional bulk RNA analyses, RNA is extracted from formalin-fixed paraffin-embedded tissues, fresh tissue or cultured cells. Strengths and weaknesses of each method are also listed.
Abbreviations: FFPE, formalin-fixed paraffin-embedded
Due to the unique workflow of scRNA-seq including single-cell isolation at the beginning and the subsequent mRNA reverse transcription from limited RNA in a single cell, multiple methods such as micromanipulation and microfluidics have been utilized and new approaches such as Smart-seq and CEL-seq are constantly being reported [24-26]. The following sections provide a brief introduction to the current strategies for single-isolation and specific protocols for scRNA-seq.
2.2 Single-cell isolation
Single-cell isolation is the first step in scRNA-seq. A brief overview of these experimental methods, along with their strengths and weaknesses, is presented in Table 1.
| Techniques | Throughput | Automation level | Impact on cell integrity | Advantages | Disadvantages | Recommended application scenario |
|---|---|---|---|---|---|---|
| Micromanipulation | Low | Manual | Gentle |
Precise picking of single cells under microscopic supervision. Fragile cells can be isolated. |
High technical requirements Time-consuming |
Precise control over individual cells is needed. |
| Laser capture microdissection (LCM) | Low | Manual | Often impairing |
Fixed tissue can be materials. Special information could be preserved. |
High technical requirements. Time-consuming. Contamination from flanking cells. |
Single cells need to be isolated from solid samples. Location of single cells is essential to know. |
| Microfluidic | High | Automatic | Diverse |
Cost-effective Low cell sample and reagent volumes. |
Low degree of flexibility offered by a specific microfluidic chip. Often restricted to one single application. |
Analyses focused on rare samples with a low number of cells. |
| Fluorescence-activated cell sorting (FACS) | High | Automatic | Often impairing |
Suitability for rare cell sorting. High flexibility in terms of cell type. |
Hard to differentiate subpopulations with similar marker expression. Loss of spatial information, cellular functions, and cell-cell interactions |
A certain cell subpopulation is needed for scRNA-seq. |
One simple approach is micromanipulation. Single cells are manually picked with a glass pipet under a microscope [5, 27], which enables sampling from a limited number of cells or fragile cells, such as early embryos [28]. Microscopic supervision of cell isolation ensures that each sample is indeed a single cell; however, this method is time-consuming and has low-throughput. Another weakness of this approach is the high technical requirements, which may cause cellular injury due to mechanical shearing [29]. Therefore, due to these obstacles, micromanipulation is now rarely being used.
Laser capture microdissection (LCM) is another approach to capture single cells from solid tissue. Under a microscope, a laser beam is focused on cells of interest and attaches these individual cells to a thin and transparent film [30]. The single cells on the film are then transferred to a microcentrifuge tube containing appropriate buffer solutions [31]. Similar to micromanipulation, LCM is also laborious and inconvenient. Another shortcoming is that this approach is technically challenging to capture the contents of a single cell cleanly, without contamination from flanking cells and without damaging the cell RNA [32]. However, LCM also has unique advantages. It enables single-cell isolation from solid samples and it can provide spatial information for the target cell, which makes it still usable in practice.
Generally, microfluidics refers to technologies using micro-scale structures to precisely control fluids at ultralow volumes, typically at the nanoliter-to-femtoliter scale [33-35]. Microfluidics platforms, such as the Fluidigm C1 microfluidic robotic platform or microdroplet-based microfluidics methods have been utilized to trap single cells [36]. On microfluidics platforms, automated single-cell isolation was followed by automated reverse transcription and preamplification-on-chip, which downscale reactions to nanoliter or picoliter volumes and further reduce costs [37]. Moreover, microdroplet-based methods have the potential to capture thousands of cells in a single experiment [38]. Although microfluidics methods have their own limitations that capture efficiency can be low for sticky cells [36], they have gained increasing popularity due to their high throughput and low analysis cost.
Fluorescence-activated cell sorting (FACS) is a specialized type of flow cytometry. First, a mixture of biological cells is combined with a specific fluorescently tagged antibody. By detecting the specific fluorescent signals from individual cells based on the tagged antibody, single cells of interest can be sorted from the heterogeneous cell populations. Due to its capability of sorting particular cells of interest and advantages of high throughput, low analysis cost, and flexibility of samples, FACS has become a widespread method for isolating thousands of single cells [29]. However, this method also has some drawbacks, including the risk of damaging cell viability during sorting and the difficulty in differentiating cell subpopulations expressing similar markers [39].
Collectively, different single-cell isolation methods have their own strengths. Micromanipulators ensure precise cell picking and minimize cell damage. LCM enables single-cell isolation from solid samples and reserve spatial information. Microfluidics methods downscale the volume of reagents which makes it cost-effective. FACS can capture particular cells of interest. Therefore, a comprehensive understanding of these single-cell separation technologies is necessary for the selection of proper single-cell isolation methods.
2.3 Specific protocols for scRNA-seq
The accuracy and sensitivity of scRNA-seq depend on its efficiency in reverse transcription and amplification processes to obtain sufficient cDNA from a limited amount of RNA in a single cell, which enables the transcriptomic analyses at a single-cell level. Thus, several specific protocols have been developed (Table 2).
| scRNA-seq | Transcript coverage | Strand specificity | UMI utility | Early multiplexing | Amplification technology |
|---|---|---|---|---|---|
| Tang method | Full length | No | No | No | PCR after polyA tailing |
| Quartz-seq | Full length | No | No | No | PCR after polyA tailing |
| Smart-seq | Full length | No | No | No | Template switching-based PCR |
| Smart-seq2 | Full length | No | No | No | Template switching-based PCR |
| STRT-Seq | 5’-end only | Yes | Yes | Yes | Template switching-based PCR |
| CEL-Seq | 3’-end only | Yes | Yes | Yes | In vitro transcription |
| MARS-Seq | 3’-end only | Yes | Yes | Yes | In vitro transcription |
| Drop-seq | 3’-end only | Yes | Yes | Yes | Template switching-based PCR |
| InDrop | 3’-end only | Yes | Yes | Yes | In vitro transcription |
| CytoSeq | Pre-defined genes only | Yes | Yes | Yes | Gene-specific primers-based PCR |
- Abbreviations: scRNA-seq, single-cell RNA-sequencing; UMI, unique molecular identifier ; PCR, polymerase chain reaction.
Protocols in scRNA-seq are characterized by different strategies used in reverse transcription reaction and cDNA amplification. Poly(A) tailing and template-switching are two main strategies in the reverse transcription reaction. “Tang method” [5] and its improved version, Quartz-seq [40], are representative methods using poly(A) tailing while Smart-seq [41], Smart-seq2 [7], STRT-seq [42] and Drop-seq [10] use template-switching to complete second-strand synthesis. cDNA amplification after reverse transcription can be achieved by either polymerase chain reaction (PCR) or in vitro transcription (IVT). PCR can be carried out after the second-strand synthesis by poly(A) tailing or template-switching. As a nonlinear amplification process, its efficiency is sequence dependent [36]. IVT is a linear amplification method that incorporates the T7 promoter in the poly(T) primers [43] ScRNA-seq protocols that apply IVT instead of PCR include CEL-seq [44], MARS-Seq [45] and inDrops [9].
Among the multiple scRNA-seq protocols mentioned above, a key difference is that some provide full-length transcript data such as Smart-seq [41] and Smart-seq2 [7], whereas others specifically count only the 3’ or 5’ ends of the transcripts such as CEL-seq [44] and MARS-seq [45]. Therefore, the selection of a specific protocol depends on the nature of the research question. Compared to methods only capturing and sequencing the 3′ or 5′ ends of the cDNAs, protocols capable of full-length transcription are more suitable for alternative splicing pattern analyses, allelic expression detection, and RNA editing identification [39]. The strength of protocols sequencing the 3′ or 5′ end of the transcript is that they are able to combine unique molecular identifiers (UMIs) [46]. These tags allow for the identification and quantification of individual transcripts, which improve the gene-level quantification and throughput [24]. Therefore, in some cases that full-length transcript data was not required, these tag-based methods have become dominant for quantification purposes, especially for larger cell populations [2].
3 APPLICATION IN BREAST CANCER RESEARCH
The rapid development of single-cell transcriptomics enables further understanding of the heterogeneity of cells in solid tumors and has great potential for clarifying the complex mechanisms of tumor development, metastasis, and drug resistance, which may provide new strategies for individualized therapeutic treatment. Additionally, multiple novel approaches beyond clustering have been used to study breast cancer when using scRNA-seq, which enables the diverse application of scRNA-seq in cancer research beyond exploring heterogeneity. This section describes studies that have used scRNA-seq to study breast cancer and discusses the promising applications of scRNA-seq in the field of individualized therapy.
3.1 Tumor heterogeneity
Gene expression profiling has been widely used to characterize bulk tumors in individual cancer patients to discover potential target therapy [47-49] but cancers usually display intratumoral heterogeneity, which may influence their therapeutic response to specific targeted therapy as well as clinical outcomes. The emergence of scRNA sequencing allows the assessment of genetic heterogeneity in breast cancer at a single-cell resolution which can help researchers to uncover the immense biological complexity in tumors.
Triple-negative breast cancer (TNBC) is a representative breast cancer subtype characterized by extensive intratumoral diversity. Although TNBC exhibits significantly higher response rates to neoadjuvant chemotherapy, compared to estrogen receptor (ER)-positive tumors, patients with TNBC who failed to achieve a pathologic complete response (pCR) were usually associated with very poor outcomes [50]. This indicated the possible existence of a minor subpopulation of TNBC cells not sensitive to traditional chemotherapy that may induce subsequent metastasis. Thus, identifying and characterizing these distinct cells could guide targeted therapy development and survival improvement in TNBC. As scRNA-seq can provide complete gene expression patterns of individual cells obscured in bulk analysis, a research group from Harvard Medical School [12] applied single-cell profiling to discover the sub-clonal heterogeneity and aggressive disease states in TNBC. By performing scRNA-seq on untreated primary TNBC tumors, the researchers confirmed the cellular heterogeneity within primary TNBCs and identified five distinct clusters of cells by clustering analyses. Among the identified five clusters of epithelial cells named as cluster 1-cluster 5, cluster 2 had the highest proportion of high-cycling cells, indicating its high proliferation ability. Further investigation showed that cluster 2 was associated with a luminal progenitor (LP) signature that was considered as the cell of origin for breast cancers [51]. The study also validated that the high expression of the cluster 2 signature was related to worse survival outcomes. The researchers applied scRNA-seq to explore tumor heterogeneity and discovered a malignant cluster subpopulation that may drive tumor progression and, thereby, lead to poor survival outcomes. These findings not only expand our knowledge on tumor heterogeneity in TNBC but also provide potential predictors or treatment targets for TNBC with poor prognosis.
Besides TNBC, scRNA-seq has also been used to comprehensively characterize heterogeneous tumors of other breast cancer subtypes. Korean researchers [13] conducted transcriptome analysis of 515 single cells from 11 patients with different breast cancer subtypes, including luminal A, luminal B, human epidermal growth factor receptor 2 (HER2), and TNBC. The most versatile subtype composition was observed for cancer cells isolated from HER2-positive tumors, from predominant HER2 to mostly TNBC type. The researchers also found that cells from the ER/HER2 double-positive tumor tended to be the ER subtype due to the low expression of HER2 module genes and were related to the corresponding predominant ER downstream signaling pathway activation. These findings suggest that molecular profiling at the single-cell level may be more detailed and accurate compared to molecular subtypes defined by bulk analyses. Based on single-cell molecular profiling, clinicians may identify ER/HER2 double-positive tumors with prominent activation of the ER downstream signaling pathway that might benefit from intensive hormone therapy, or identify HER2-positive tumors with low levels of HER signaling pathway activation but higher basal gene expression that might be resistant to anti-HER2 target therapy. Guided by single-cell molecular profiling, clinicians could choose more effective therapeutic regimen and improve the patients’ prognosis.
Single-cell profiling of circulating tumor cells (CTCs) can also provide unique insights into tumor heterogeneity. Genotypic and phenotypic characterization of CTCs has the potential to provide a better understanding of tumor evolution and identify metastasis-initiating cells [52-55] to elucidate the onset of metastasis and identify potential therapeutic targets that could be used to prevent or treat metastasis. A recent singlecell study on CTCs isolated from blood samples of patients with primary and metastatic breast cancer [14] reported that individual CTCs had significant heterogeneity and identified two distinct clusters of CTCs named as Cluster I and Cluster II. Cluster I was characterized by the expression of genes associated with the epithelial-mesenchymal transition while Cluster II generally showed low to undetectable expression of these genes, which indicated that Cluster I may be related to cancer progression and poor prognosis. The researchers also compared the profiles of CTCs and breast cancer cell lines that are widely used in drug discovery. Significant differences between CTCs and cell lines were identified in the expression of certain genes, levels of growth factors and clinically informative phenotypes, which indicated that traditional cell lines may not perfectly simulate the seeding process of metastasis by CTCs and that conventional therapies designed and tested based on these cell lines may be inadequate for eliminating metastasis. Thus, appropriate experimental systems for drug discovery should be carefully selected with the help of cell-to-cell profiling of CTCs. Meanwhile, liquid biopsies may help to identify patients with specific CTC clusters and inform the selection of therapeutic interventions targeting overexpressed gene products and activated pathways in these CTC clusters.
3.2 Tumor microenvironment
The tumor microenvironment comprises of heterogeneous cellular populations including tumor cells and the surrounding nonmalignant cells, such as vascular, immune, and fibroblast cell types [56]. As previously described [57-59], constant cross-talk between tumor cells and the surrounding microenvironment was associated with treatment response and survival outcomes of malignancies. ScRNA-seq is a powerful tool to explore the complex tumor microenvironment and further facilitate individualized therapy and overcome drug resistance. Here, we briefly introduce a few key studies applying scRNA-seq methods to explore tumor environment heterogeneity.
Researchers from the Memorial Sloan Kettering Cancer Center conducted a single-cell analysis of the immune environment of eight primary breast carcinomas composed of one HER2-positive tumor, two TNBCs and five ER-positive tumors, and found a large degree of diversity in the immune composition of each tumor [60]. By assessing the immune cells captured in the normal and malignant breast tissues, lymph nodes, and peripheral blood, the researchers found that the immune phenotype was associated with the tissue of residence, suggesting that biomarkers based on blood samples may not reflect the immune phenotype states in tumors [15]. In-depth analyses of T cells, which are considered more clinically relevant, revealed that intratumoral T cells displayed continuous activation and differentiation transitions rather than a few discrete and stable cell states. Therefore, the traditional classification of T cells in tumors may underestimate the complexity of the T cell populations. The authors of this study suggested that the single-cell profiling of diverse immune cells facilitated the understanding of complex mechanisms in immune enhancement and suppression in the tumor environment, and the co-expression of checkpoint receptor genes identified in some regulatory T cells clusters may be potential immunotherapy targets.
Another single-cell profiling study of T cells in the breast cancer microenvironment conducted by a research group in Australia analyzed data from 6,311 T cells isolated from human breast cancer tissue. Their analyses revealed heterogeneity in the CD8+ population [16]. CD8+ T cells with features of tissue-resident memory T (TRM) cell differentiation were found to express both immune checkpoints and cytotoxic effector proteins. The expression of inhibitory receptor genes such as PDCD1 and CTLA4 was higher in these distinct TRM cells. Thus, CD8+ TRM cells may be associated with immunosurveillance and could be potential immunotherapy targets. As indicated in that study, scRNA-seq enabled the discovery of minor subgroups of immune cells that were related to immunosuppression or immunosurveillance. Biomarkers of these distinct immune cells may serve as prognostic factors or therapeutic targets, allowing individual risk assessment and stratification of patients for targeted therapies.
In summary, scRNA-seq helped to explore the complexity of cell populations in the breast cancer microenvironment and revealed distinct cell subpopulations with the potential to become individualized immunotherapy targets.
3.3 Diverse approaches beyond exploring heterogeneity
As mentioned above, many cancer studies have explored the cellular heterogeneity within a tumor. However, scRNA-seq can also be applied to many other types of breast cancer research beyond exploring heterogeneity. For instance, scRNA-seq has been used to study the cell-to-cell communications mediated by secreted factors and its role in tumor progression [18]. The different expression levels of receptors and their ligands between tumoral and normal tissues, and the correlation between the expressions of these specific ligand-receptor pairs could serve as indicators of cell-cell communication. By providing information on the expression level of ligands and receptors of individual cells, which determines “who talks to whom”, single-cell transcriptome data has enabled researchers to map a more-defined cell-cell communication network with distributions of receiving and signaling cells in distinct cell populations. Moreover, with increased knowledge of the heterogeneity of the tumor parenchyma, research on the communications between distinct cell populations have also gained attention. ScRNA-seq could integrate the ligand-receptor analysis and clonal analyses to construct a heterotypic cell-cell communication network, and further facilitate the study on cell-to-cell communications between cell populations of clinical significance, which might help to find novel targets of individualized therapy.
Besides genetic heterogeneity, tumors also display regulatory heterogeneity [61]. While the current scRNA-seq is a powerful tool to study heterogeneity in lineage, it is still difficult to reliably measure tumor-cell regulatory heterogeneities. Thus, complementary approaches are needed to profile single-cell regulatory states. One example is the in situ 10-cell RNA-seq (10cRNA-seq) [20], which combines the single-cell resolution of laser capture and improves the preamplification procedure; enabling RNA-seq of 10 micro-dissected cells and making it a reliable, unbiased, and sensitive method to measure cell-state heterogeneity in tumors.
Researchers have also used scRNA-seq to identify progenitor or stem-like cells. By applying a marker-free system biology approach called LandSCENT in single-cell transcriptome data from the human mammary epithelium, researchers have identified a novel bipotent stem-like state that was also correlated with poor prognosis in basal breast cancer [19]. The novel marker-free computational approach used in this study allowed the estimation of cell potency to enable the identification of rare cell populations representing progenitor or stem-like cells. Its future application in other tumor types to identify putative cancer stem cells warrants investigation.
With the increasing awareness of the importance of special cellular information in cancer research, novel computational approaches have been developed to reconstruct the spatial organization of cells lost in the process of scRNA-seq analysis. Researchers in Sweden applied spatial transcriptomics technology to identify immune cell distribution in HER2+ breast cancer [21]. They constructed and analyzed three-dimensional images of the transcriptional landscape to reveal immune cell infiltration patterns among different patients, which may provide an opportunity for the design of personalized treatments.
3.4 Therapeutic selection and monitoring
The continued discovery of genotypically and phenotypically distinct cell subpopulations has changed our view of cancer from a group of homogenous tumor cells to a constellation of heterogeneous, continuously evolving cancer subpopulations [14]. ScRNA-seq method can be an important tool to identify optimum combined therapies to effectively target these heterogeneous cell populations. In addition, scRNA-seq may identify alterations associated with therapeutic resistance in distinct cell clusters to support individualized cancer treatment.
To identify cell diversity by transcriptome profiling, scRNA-seq was utilized to explore possible predictors of clinical outcomes and drug targets in breast cancer by characterizing cell subsets with metastatic potential. For example, transcriptome profiling of migratory breast cancer cells using scRNA-seq by researchers at the University of Michigan [62] revealed that the over-expression of certain genes in migratory cells was associated with poor prognosis in breast cancer patients, suggesting that these regulators of cell migration may be potential prognostic markers or drug targets. Similarly, researchers at the University of California conducting scRNA-seq in matched primary breast cancer and micrometastases discovered a distinct transcriptome program related to poor prognosis in micrometastatic cells and identified mitochondrial oxidative phosphorylation (OXPHOS) as a top pathway upregulated during metastatic seeding [63]. Selective inhibition of OXPHOS may be a novel target therapy to prevent breast cancer metastasis. Ongoing research is expected to discover additional factors for more accurate prediction of prognosis and novel drug targets to facilitate pharmaceutical research, which may dramatically affect the current treatment of patients with breast cancer.
Currently, gene expression signatures are mainly generated by conventional molecular profiling methods such as micro-array or bulk RNA-seq, which measures the average values of gene expression. scRNA-seq enables profiling at the single-cell level and detection of gene expression differences within cell populations. Therefore, novel biomarkers related to cancer progression and response to target therapies might be revealed by scRNA-seq. By comparing single-cell transcriptome profiles between trastuzumab-treated and non-treated patients with HER2+ breast cancer, researchers found that trastuzumab-enhanced matrix Gla protein (MGP) gene expression was associated with a better prognosis [64]. Moreover, they constructed a 48-gene expression signature that was associated with cardiomyocyte death, which could serve as a predictor for trastuzumab-mediated cardiotoxicity. Although no other gene expression signature-based on scRNA-seq data has yet been reported, additional novel signatures will likely be discovered with increased application of scRNA-seq. These signatures may better guide therapeutic decision-making in clinical practice.
As a major obstacle in the treatment of breast cancer, drug resistance is related to tumor heterogeneity. Therefore, the detection of cell populations able to survive anticancer treatment is critical to the prediction and reversion of drug resistance. The discovery of minor groups of drug-resistant cells by genetic studies performed on bulk samples was difficult. However, the development of scRNA-seq enables extensive single-cell gene expression profiling to characterize these drug-resistant cells and identify the underlying mechanism of drug resistance acquisition. Researchers have performed scRNA-seq in docetaxel-resistant cells derived from the luminal-type breast cancer cell line MCF7 [65]. Epithelial-to-mesenchymal transition and stemness-related genes were found upregulated, while cell-cycle-related genes were found downregulated in these chemo-resistant cells. A similar gene-expression pattern was also identified in a subgroup of untreated cells, suggesting the existence of cell subpopulations with an inherent predisposition toward docetaxel resistance. Identification of distinctive gene expression patterns of cells not sensitive to specific drugs may inform the design of multi-gene panels to predict the efficacy of specific drugs and assist clinicians in choosing an individualized therapeutic regimen.
In metastatic breast cancer research, the significance of CTCs in selecting appropriate therapies, monitoring therapeutic response, and innovating new treatments has been widely recognized [66]. Multiple nucleic acid- and protein-based assays have been developed to assess the ER and HER2 status in CTCs, which help clinicians to identify candidates for endocrine therapy and anti-HER2 therapy [67, 68]. Researchers also found that other biomarkers beyond ER and HER2 in CTCs could guide treatment decisions and monitor treatment efficacy as well. For instance, CD133 expression in CTCs may serve as a potential marker for chemoresistance [69] while CTC-Endocrine Therapy Index might predict resistance to endocrine therapy in MBC patients [70]. The heterogeneity and rarity of CTCs warrant the use of single-cell technologies to provide us with a more comprehensive understanding of CTCs. In a recent study, the molecular analysis of single CTCs isolated from metastatic breast cancer patients has been achieved in a noninvasive way and researchers have identified the heterogeneity of PIK3CA mutational status at single-cell level in CTCs isolated from individual patients [71]. Thus, similar studies could be conducted to analyze other clinically relevant genetic mutations in CTCs. CTC-derived preclinical models could then be designed to develop new drugs targeting specific genetic mutations. Furthermore, corresponding clinical trials can be designed to investigate the prognostic value of gene mutations detected in CTCs and monitor the efficacy of newly developed drugs, which may help clinicians to more effectively tailor therapeutic regimens for individual patients.
ScRNA-seq is a powerful tool in discovering novel therapies associated with tumor mutation burden or immune checkpoint crosstalk. A Korean research group analyzed scRNA-seq data from breast cancer and immune cells to explore potential approaches for improving the therapeutic outcome of immune checkpoint inhibition. The study mainly analyzed the tumor mutation burden (TMB), immune checkpoint crosstalk, radiosensitivity, and their relationships to test the hypothesis that the radiosensitivity of tumor cells might be related to its programmed death-ligand 1 (PD-L1) expression or TMB [22]. According to their results, radioresistant tumor cells were associated with a higher rate of PD-L1 positivity and TMB. Additionally, increased immune checkpoint crosstalk between cancer cells and immune cells was observed in TNBC compared to those in the luminal and HER2-positive subtypes. These findings suggest that radiotherapy could be combined with immune checkpoint blockades to improve the outcome of tumors with radiosensitive cells characterized by high PD-L1 expression and higher TMB in TNBC. In another recent study that performed scRNA-seq analysis in TNBC murine models, a high mutation burden was also found associated with immune checkpoint therapy response [72]. In these models, B cell activation of T cells was identified as an important mechanism that mediated response to checkpoint inhibitors, which indicated that the combined novel drugs that could induce B cell-dependent T cell activation may enhance the anti-tumor response of immune checkpoint therapy [72]. Single-cell profiling also helped researchers to find a distinct immunosuppressive immature myeloid cell (IMC) population that infiltrated the fast-evolving cyclin-dependent kinases 4 and 6 (CDK4/6) inhibitor-resistant tumors [73]. According to this study, combinatorial immunotherapy of IMC-targeting tyrosine kinase inhibitor cabozantinib and immune checkpoint blockade could enhance anti-tumor immunity and overcome CDK4/6 inhibitor resistance [73]. Single-cell profiling showed its superiority in these studies to identify potential biomarkers and optimal combination strategies of immune therapy in the management of breast cancer.
ScRNA-seq was also performed to study the synergistic effect of the anti-tumor immune response induced by chemotherapy in breast cancer. A recent study conducted by Chinese researchers identified the phenotype switch of B cells induced by chemotherapy in breast cancer patients and further explored the underlying mechanism. scRNA-seq of tumor-infiltrating B cells was performed in paired clinical samples of pre- and post-neoadjuvant chemotherapy collected from breast cancer patients [23]. The results showed that a distinct B cell subset that expressed high levels of inducible T-cell co-stimulator ligand (ICOSL) significantly increased after neoadjuvant chemotherapy. Compared to patients with stable diseases or progression, the ICOSL+ B cell density in the residual tumor was markedly higher in patients with partial or complete remission (P < 0.001), indicating that the ICOSL+ B cell subset was also related to improved therapeutic efficacy. Meanwhile, survival analyses indicated that ICOSL+ B cell abundance was an independent positive prognostic factor for disease-free survival (hazard ratio [HR], 0.275; 95% confidential interval[CI], 0.134-0.564) and overall survival (HR, 0.232; 95% CI, 0.090-0.601). The underlying mechanism behind the B cell switch was studied in mouse models. The researchers found that the anti-tumor immune response was elicited by these distinct B cell subsets after chemotherapy, which enhanced therapeutic efficacy. Further analyses found the generation of ICOSL+B cells depended on complement receptor type 2 (CR2) signaling, whereas tumoral CD55 expression could inhibit this complement-dependent ICOSL+ B cell induction and thereby undermine anti-tumor immunity. Therefore, CD55 could be a potential target to enhance immune response and facilitate immunogenic cell death.
The combination of scRNA-seq with other technologies has further facilitated the development of personalized treatment. Combined with a scalable hydrodynamic scRNA-seq barcoding technique, a high-throughput contamination-free scRNA-seq, Hydro-Seq, enables the analysis of small numbers of CTCs in 10 mL of blood [74]. Hydro-Seq allowed the detection of ER, progesterone receptor (PR), HER2 expression, and molecular profiles in CTCs, which could be discordant compared to primary tumors. Identification of the clinical markers of CTC in lipid biopsy may help to identify drug targets for personalized therapy. Transcriptome analysis of CTCs also increases our understanding on the intra-tumor cellular heterogeneity and evolution of tumors following therapy, which could provide insights into monitoring target therapeutics and processes underlying tumor metastasis. Using functional cellular approaches combined with scRNA-seq, researchers have analyzed cells with different degrees of cancer stem cells (CSCs) properties in breast cancer cell lines and identified potential breast cancer biomarkers related to CSC properties [75]. Due to their self-renewal capacity and potential to differentiate into cancer cells, CSCs have been associated with tumor progression, metastasis, and treatment resistance, leading to poor prognosis [76]. Many useful markers for the identification of CSCs can also serve as therapeutic targets to eliminate CSCs, especially those involved in pathways associated with self-renewal and epithelial–mesenchymal transition (EMT), such as Notch, Hedgehog, and Wnt signaling pathways [77]. The inhibition of the Notch signaling can be achieved by γ-secretase inhibitors [78] and clinical trials have already been launched to evaluate the efficacy of γ-secretase inhibitor MK-0752 (Merck) in metastatic breast cancer [79]. Multiple CSC-based targets have been investigated to inhibit Hedgehog and Wnt signaling as well, such as vismodegib [80] and vantictumab [81]. Therefore, the identification of CSC markers and novel drugs targeting CSCs may provide new opportunities for individualized therapy. A newly developed method has enabled the detection of cell-level mutations from scRNA-seq data [82], which further broaden the application of scRNA-seq in future cancer research. Specific somatic alternations were found associated with drug responses in breast cancer. For example, GATA3 mutation may predict better response for aromatase inhibition [83] and PI3K and/or ERBB2 mutations may indicate the sensitivity of HER2+ tumors to neoadjuvant docetaxel, carboplatin, trastuzumab and lapatinib regimen [84]. Mutation profiling based on scRNA-seq in breast cancer research has not been conducted yet. However, with the development of new methods like the one mentioned above, it is believed that scRNA-seq will be used to identify cell-specific mutations in future studies and help to predict individual drug-response.
Individualized cancer management largely depends on the precise detection of specific molecular subtypes or therapeutic targets. Although heterogeneity-based personalized treatment has progressed substantially in breast cancer, with a growing list of target agents in adjuvant, neoadjuvant, and metastatic settings, there is an unmet need to enhance precise medicine to improve survival and eliminate late relapse. As mentioned above, scRNA-seq is a powerful tool in exploring tumor and tumor environment heterogeneity. The detection of distinct cell subsets not sensitive to conventional therapy may explain intrinsic drug resistance and targeted agents against these cell subsets could provide crucial clinical benefits. By using scRNA-seq, researchers have found that the endocrine-resistance of fulvestrant and tamoxifen was caused by a group of pre-existing genetically distinct cells not sensitive to endocrine therapy and these cells were highly selected during treatment [85]. Therefore, the innovation of novel drugs targeting these cells might reverse the endocrine resistance. ScRNA-seq could also be used to detect the emergence of resistant cell subsets after treatment, which could have significant clinical implications for second-line treatment decision-making on available or new target drugs. For instance, in a translational research mentioned above, researchers have used scRNA-seq to trace the tumor evolution during the combination treatment of anti-HER2/neu antibody and CDK4/6 inhibitor and discovered that the acquired resistance was related to infiltrating immunosuppressive immature myeloid cell (IMC) population [73]. They also found that combinatorial immunotherapy of IMC-targeting drugs and immune checkpoint blockade was an effective regimen to treat these fast-evolving CDK4/6 inhibitor-resistant tumors [73]. Collectively, integrating scRNA-seq into basic and translational research could promote personalized therapy by identifying potential treatment targets to develop novel drugs and reveal promising biomarkers to monitor treatment efficacy and guide therapeutic decision-making.
4 FUTURE OUTLOOK
ScRNA-seq is a promising new technology that provides a transcriptomic analysis of individual cells and is a powerful tool to address the inherent complexity of breast cancer and its tumor environment to pave the way for individualized therapy. However, as a recently developed technology, scRNA-seq still has some limitations.
First, both cell integrity and cell viability are essential for subsequent single-cell analyses. It means that single cells need to be isolated from each other quickly and accurately with minimum damage to the cells during the single-cell isolation process. However, mechanical dissociation of tissue might injure cell integrity and enzymatic treatment using trypsin, collagenase, and/or papain to isolate single cells from tissues may affect cell viability or lead to transcriptional changes. Thus, there is a need to improve approaches to enable the efficient “gentle” extraction and capture of living cells and avoid the potential damage to single cells caused by enzymatic treatment. Another limitation is the relatively high cost of single-cell sequencing. Even though the currently available systems have cut down the price of sequencing per cell to a seemingly acceptable level, the combined cost is still too high because tens of thousands of cells must be analyzed in some instances. Reduced sequencing costs would facilitate the widespread use of scRNA-seq in tumor research. Furthermore, integrating scRNA-seq with other single-cell genomic or protein information is also a future experimental challenge. The combination of scRNA-seq and DNA sequencing may not only provide us with more information on the interaction between the epigenome and transcriptome, but also provide insight into the transcriptomic phenotype of cells that share somatic mutations in DNA. Other new directions of technology development include imaging a cell before sequencing to acquire morphologic information and localizing the cell in a tissue or microenvironmental niche to acquire spatial information. In addition to technological improvements, computational methods also need to be upgraded to separate the technical noise. Failure to account for technical noise in scRNA-seq data can lead to biased downstream analyses and misleading results, therefore it has become a key challenge to accurately separate the technical noise from biological heterogeneity [86]. In order to take full advantage of scRNA-seq and better quantify biological variation, advanced methods for data analysis are needed. These limitations are anticipated to be addressed by rapid scientific and technological development. The application of scRNA-seq to tumor biology research could further expand our understanding on tumor heterogeneity, provide insights into molecular mechanisms in tumor evolution and metastasis, and facilitate the discovery of novel therapeutics for more prosperous individualized cancer management
5 CONCLUSIONS
Single-cell sequencing has provided enlightening insights into the study of tumor heterogeneity and has allowed a more comprehensive understanding of tumor development and progression, which are crucial for the development of targeted therapy and the realization of individual management. Widespread use of scRNA-seq could lead to a profound revolution in our understanding of breast cancer and facilitate the discovery of more effective therapeutic methods to prevent cancer relapse and improve survival benefit.
DECLARATIONS
AUTHORSHIP
Conception and design: all authors. Manuscript writing: SND. Manuscript revision: XSC and KWS. All authors read and approved the final manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
FUNDING
The authors received financial support from the National Natural Science Foundation of China (Grant Number: 81772797), Shanghai Municipal Education Commission—Gaofeng Clinical Medicine Grant Support (20172007); Ruijin Hospital, Shanghai Jiao Tong University School of Medicine “Guangci Excellent Youth Training Program” (GCQN-2017-A18). All these financial sponsors had no role in the study design, data collection, analysis, or interpretation.
Open Research
DATA AVAILABILITY STATEMENT
Not applicable.




