Association between glioblastoma cell-derived vessels and poor prognosis of the patients
Abstract
Background
Vessels with different microcirculation patterns are required for glioblastoma (GBM) growth. However, details of the microcirculation patterns in GBM remain unclear. Here, we examined the microcirculation patterns of GBM and analyzed their roles in patient prognosis together with two well-known GMB prognosis factors (O6-methylguanine DNA methyltransferase [MGMT] promoter methylation status and isocitrate dehydrogenase [IDH] mutations).
Methods
Eighty GBM clinical specimens were collected from patients diagnosed between January 2000 and December 2012. The microcirculation patterns, including endothelium-dependent vessels (EDVs), extracellular matrix-dependent vessels (ECMDVs), GBM cell-derived vessels (GDVs), and mosaic vessels (MVs), were evaluated by immunohistochemistry (IHC) and immunofluorescence (IF) staining in both GBM clinical specimens and xenograft tissues. Vascular density assessments and three-dimensional reconstruction were performed. MGMT promoter methylation status was determined by methylation-specific PCR, and IDH1/2 mutations were detected by Sanger sequencing. The relationship between the microcirculation patterns and patient prognosis was analyzed by Kaplan-Meier method.
Results
All 4 microcirculation patterns were observed in both GBM clinical specimens and xenograft tissues. EDVs were detected in all tissue samples, while the other three patterns were observed in a small number of tissue samples (ECMDVs in 27.5%, GDVs in 43.8%, and MVs in 52.5% tissue samples). GDV-positive patients had a median survival of 9.56 months versus 13.60 months for GDV-negative patients (P = 0.015). In MGMT promoter-methylated cohort, GDV-positive patients had a median survival of 6.76 months versus 14.23 months for GDV-negative patients (P = 0.022).
Conclusion
GDVs might be a negative predictor for the survival of GBM patients, even in those with MGMT promoter methylation.
Abbreviations
-
- 3D
-
- three-dimensional
-
- bFGF
-
- basic fibroblast growth factor
-
- CI
-
- confidence interval
-
- DAB
-
- 3,3ʹ-diaminobenzidine
-
- DAPI
-
- 4′,6-diamidino-2-phenylindole
-
- ECMDV
-
- extracellular matrix-dependent vessel
-
- EDTA
-
- ethylenediaminetetraacetic acid
-
- EDV
-
- endothelium-dependent vessel
-
- EGF
-
- epidermal growth factor
-
- FFPE
-
- formalin-fixed, paraffin-embedded
-
- GBM
-
- glioblastoma
-
- GDEC
-
- GBM-derived endothelial cell
-
- GDV
-
- GBM cell-derived vessel
-
- HR
-
- hazard ratio
-
- IDH
-
- isocitrate dehydrogenase
-
- IF
-
- immunofluorescence
-
- IHC
-
- immunohistochemistry
-
- KPS
-
- Karnofsky Performance Status
-
- LNA
-
- Locked Nucleic Acid
-
- MGMT
-
- O6-methylguanine DNA methyltransferase
-
- MSP
-
- methylation-specific PCR
-
- MV
-
- mosaic vessel
-
- MVD
-
- microvessel density
-
- PAS
-
- periodic acid-Schiff
-
- PBL
-
- peripheral blood lymphocyte
-
- PBS
-
- phosphate-buffered saline
-
- ROI
-
- regions of interest
-
- Sox2
-
- sex determining region Y-box 2
-
- VEGF
-
- vascular endothelial growth factor
-
- VM
-
- vasculogenic mimicry
-
- WHO
-
- World Health Organization
1 BACKGROUND
Glioblastoma (GBM) is the most common malignant primary brain tumor [1]. Patients with GBM survive an average of 14 months after diagnosis, even after the administration of standard treatment, including surgery, radiotherapy, and chemotherapy [1]. Effective strategies, including anti-angiogenic therapy which targets the enriched blood supply of GBM, have been studied intensively. Bevacizumab, an antibody against vascular endothelial growth factor (VEGF), has been studied in patients with recurrent and progressive GBM [2]. However, compared with standard treatment alone, bevacizumab combination therapy failed to show any improvement in overall survival for patients with newly diagnosed GBM [3]. It remains unclear why bevacizumab does not work well in patients with GBM, but has resulted in dramatic improvements in patients with some other cancer types, such as colon cancer [4] and rectal cancer [5].
In 1999, a novel vascular type, vasculogenic mimicry (VM), was first reported in melanoma [6]. VM describes the two types of vessels to complement endothelium-dependent vessels (EDVs), which are extracellular matrix-dependent vessels (ECMDVs) and tumor cell-derived vessels. Our group first reported the presence of VM in gliomas in 2005 [7]. The physiological connection between the endothelial-lined vasculature and VM channels was then reported later [8]. We further found that patients with VM-positive glioma survived a shorter period of time than patients with VM-negative glioma [9]. VM was then confirmed as an adverse prognostic factor in newly diagnosed GBM [10] and breast cancer [11, 12]. It may represent an important tumor survival mechanism contributing to the failure of current anti-angiogenic therapy, which aims to completely deprive the tumors of their blood supply [13].
A better understanding of vascularization is necessary to improve the efficiency of anti-angiogenic therapy against GBM. Recent studies have shown that EDVs are not the exclusive means of blood supply to tumors [14, 15]. In addition to the well-studied angiogenesis, several new microcirculation patterns have been revealed, including mosaic vessels (MVs) derived from both endothelial and tumor cells [16], and VM channels [17], by which tumor tissues nourish themselves. In our previous study, we identified GBM-derived endothelial cells (GDECs) and further investigated the vessel formed by GDECs (we named it “GBM cell-derived vessel [GDV]”) [18]. In the present study, we examined the microcirculation patterns of GBM and analyzed their roles in patient prognosis together with the well-known GMB prognosis factors, such as O6-methylguanine DNA methyltransferase (MGMT) promoter methylation status and isocitrate dehydrogenase (IDH) mutations.
2 MATERIALS AND METHODS
2.1 Patients and clinical specimens
We performed a retrospective study of 80 patients with GBM treated at Sun Yat-sen University Cancer Center (SYSUCC, Guangzhou, Guangdong, China) between January 2000 and December 2012. All included patients were pathologically confirmed as GBM according to the 2016 World Health Organization (WHO) classification for central nervous system tumors. All patients were treatment-naive before surgery.
The formalin-fixed, paraffin-embedded (FFPE) tissue specimens of GBM from the selected patients were retrieved from the Department of Pathology at SYSUCC. Tumor sections were reviewed by two neuropathologists to verify the GBM diagnosis. This study was approved by the Ethics Committee of SYSUCC (case number: GZR2013-052). All patients provided written informed consent.
2.2 Mice
Four-week-old nude mice weighing 16 to 20 g were obtained from the Medical Experimental Animal Center of Guangdong Province (Guangzhou, Guangdong, China). All nude mice were housed under standard conditions and cared for according to the institutional guidelines for animal care. All animal procedures were performed under the ethical standards of the Accreditation of Laboratory Animal Care and approved by the Ethics Committee of SYSUCC.
2.3 Cell culture
The glioma stem-like cells (GSC-1), stably expressing CD133 and sex determining region Y-box 2 (Sox2), were generated from tumor tissue of a patient with GBM(18) and cultured in Dulbecco's Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F12; Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 20µL/mL B27 (Thermo Fisher Scientific), 20 ng/mL epidermal growth factor (EGF; Peprotech, Rocky Hill, NJ, USA), 20 ng/mL basic fibroblast growth factor (bFGF; Peprotech), and 1% penicillin/streptomycin (Thermo Fisher Scientific, Waltham, MA, USA) at 37°C in 5% CO2.
2.4 Xenograft specimens
A total of 1 × 106 GSC-1 cells in 200 µL serum-free medium were inoculated subcutaneously into the flanks of the nude mice. Eleven days after the inoculation, a tumor mass was observed. All nude mice were euthanized 18 days after inoculation, and the xenograft tumor tissues were collected, fixed with 10% formalin, and embedded in paraffin for further experiments.
2.5 Immunohistochemistry (IHC)
Standard streptavidin-biotin-peroidase complex method was used for IHC staining. FFPE sections were stained with an antibody against CD34 (1:1000; ab198395, Abcam, Boston, MA, USA) and periodic acid-Schiff (PAS; BD, Franklin Lakes, NJ, USA), as previously described [7]. Briefly, FFPE sections were deparaffinized by xylene and absolute ethanol, rinsed in distilled water and exposed to 3% H2O2 for 10 min. Slides were put in ethylenediaminetetraacetic acid (EDTA) antigen-unmasking solution heating in oven at 90°C for 15 min and then cooled to room temperature in unmasking solution. Slides were then washed in phosphate-buffered saline (PBS) and incubated with the primary antibody overnight at 4°C. After being washed in PBS, the slides were incubated with the secondary antibody (1:1000; Golden Bridge, Beijing, China). The nuclear expression of CD34 was visualized with chromogen 3,3ʹ-diaminobenzidine (DAB; Golden Bridge). Slides were then rinsed with distilled water for 5 min and incubated with PAS stain for 15 min, and the cell nuclei were stained with hematoxylin.
2.6 Immunofluorescence (IF) staining
The deparaffinization, rehydration, and antigen retrieval of tumor sections were as described in IHC. Slides were washed with PBS before incubation with the first primary antibody, rabbit monoclonal anti-CD34 (1:1000; ab198395, Abcam), at 4°C overnight. After being washed in PBS, the sections were incubated with mouse monoclonal anti-GFAP (1:1000; ab10062, Abcam) or anti-laminin antibodies (1:1000; ab7463, Abcam). Next, slides were washed with PBS and incubated with Alexa Fluor® 488-conjugated goat anti-rabbit IgG (1:1000; O-11038; Life Technologies, Carlsbad, CA, USA) and Alexa Fluor® 594-conjugated goat anti-mouse IgG (1:1000; AK-A11005; Life Technologies) for 40 min at room temperature. Nuclear staining was performed with 4′,6-diamidino-2-phenylindole (DAPI; D3571; Life Technologies), and the slides were then mounted with Slow Fade Gold antifade reagent (S36938, Thermo Fisher Scientific).
2.7 Observation and density assessment of microcirculation patterns
The IHC- and IF-stained slides were reviewed by Xin Mei and Yin-Sheng Chen using optical microscope (Olympus Corporation, Shinjuku,Tokyo, Japan) for IHC and confocal laser microscope (Olympus Corporation) for IF following the methods described by Folberg et al. [19]. EDV had a CD34-positive endothelium wall. ECMDV had extracellular matrix surrounding the lumen but without CD34 or GFAP staining. Vessels with only a GBM cell lining were regarded as GDVs, and vessels comprising a mixture of tumor cells and endothelial cells were regarded as MVs (Supplementary Table S1). Vessel density was assessed as described by Paschoal et al. [20]. Briefly, each IHC- or IF-stained section was first observed at low optical power to choose two regions of interest (ROI). The number of vessels expressing the biomarkers was then counted at high magnification (high optical power, 400 ×) within the ROI. Next, the mean microvessel density (MVD) was calculated.
2.8 Three-dimensional (3D) reconstruction of microcirculation patterns
We reconstructed the microcirculation patterns in human GBM tissue sections (1 mm thickness) with IF staining for CD34 and GFAP and analyzed it using Fluoview 3.0 software (Olympus Corporation). Images were stored in TIFF format. One 3D reconstruction picture comprised more than twenty image frames, and the thickness of each layer was 2 µm. The visualization of the 3D shapes of the cell nuclei (blue), endothelial vessels (green), and GBM cells (red) was achieved by representing the traces of these objects using “Reconstruct”. The 3D models were saved as a 360° image series and then exported into Fluoview 3.0 software, where the files were compressed (JPEG).
2.9 Methylation-specific PCR (MSP) of MGMT
Sufficient tissue was available from 61 GBM patients to determine the methylation status of the MGMT promoter region using MSP. Genomic DNA was extracted from paraffin-embedded specimens according to the manufacturer's instructions (TianGen DNA Mini Kit, Beijing, China). The methylation status was determined by performing the bisulfite modification, which converts unmethylated but not methylated cytosines to uracils. The genomic DNA (50 ng) was amplified using primers designed to detect the methylated or unmethylated sequences by MSP. MSP was performed according to the manufacturer's protocol (EZ DNA Methylation Gold kit, Zymo Research, Irvine, CA, USA). The primer sequences were as follows: forward primer, 5′-GTTATGAATGTAGGAGCCCTTATG-3′; reverse primer, 5′-TGACAACGGGAATGAAGTAATG-3′, which were specific for either the methylated or modified unmethylated DNA [21]. DNA obtained from normal peripheral blood lymphocytes (PBLs) served as the negative control, and enzymatically methylated DNA from PBL was used as the positive control. Ten microliters of each 50-µL MSP product were directly loaded onto nondenaturing 6% polyacrylamide gels, stained with ethidium bromide, and examined under ultraviolet illumination.
2.10 Detection of IDH1/2 mutations
Bidirectional Sanger sequencing (Qiagen, Hilden, Germany) was performed to detect the mutations of IDH1/2. The primers for IDH1 (129 bp for codon 132) and IDH2 (290 bp for codon 172) were listed here: IDH1-F-5′-CGGTCTTCAGAGAAGCCATT-3′, IDH1-R-5′-ATTCTTATCTTTTGGTATCTACACC-3′, IDH2-F-5′- GGGGTTCAAATTCTGGTTGA-3′, IDH2-R-5′-ACTGGAGCTCCTCGCCTAG-3′.
Briefly, PCR was performed in a total volume of 60 µL using 15 ng of DNA and Taq Polymerase Platinum in standard PCR buffer (BioRad Laboratories, Pleasanton, CA, USA). An initial denaturation step at 94°C for 2 min was followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 68°C for 30 s. PCR products were purified using the QIA quick 96 PCR Purification Kit (28181; Qiagen). The purified PCR products (1.5 µL) were sequenced using Applied Biosystems Big Dye 3.1 dideoxy chain termination sequencing chemistry in conjunction with fluorescent-based capillary sequencers (Model 3730xl, Applied Biosystems Division of Perkin Elmer, Foster City, CA, USA) for electrophoresis, data collection, and base calling.
Locked Nucleic Acid (LNA) Sanger sequencing was performed in a separate reaction by amplifying the mutation hotspot regions using the pyrosequencing PCR primers and LNA (TAGGTCGTCA/3InvdT/) at a final concentration of 70 nmol/L. The PCR product was sequenced using the Big Dye Terminator v1.1 Sequencing Kit (Applied Biosystems; Foster City, CA, USA).
2.11 Statistical analysis
The constituent ratios of the microcirculation patterns were separately calculated as the mean value of each sample stained with IHC and IF. The microcirculaiton pattern was determined by the staining results from either IHC or IF. We then compared the prognosis of patients with different microcirculation patterns. The overall survival was defined from the date of diagnosis to the date of the death or to the last observation date (December 31, 2018). Multivariable analysis was performed including gender, age and MGMT promoter methylation status for the prognosis of GDVs. The hazard ratios and 95% confidence intervals for patient outcome were estimated by univariate and multivariate Cox regression. Kaplan-Meier survival curves were generated for each set. Statistical analyses were performed using SPSS version 19 (SPSS Inc., Chicago, IL, USA), and P values less than 0.05 were considered significant.
3 RESULTS
3.1 Microcirculation patterns in GBM
Four microcirculation patterns, EDV, ECMDV, GDV, and MV, were assessed in both clinical GBM and xenograft specimens by IHC and IF staining. Among them, EDVs existed in all tissue specimens. However, ECMDVs, GDVs, and MVs were only observed in 27.5%, 43.8%, 52.5% of specimens from GBM patients and 40.0%, 5.0%, 70.0% of the specimens from xenografts, respectively.
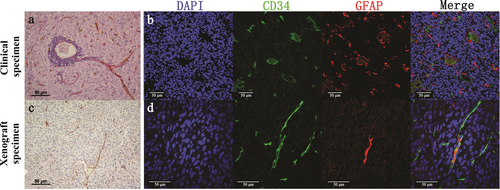
Most of the intratumoral vessels (90.2%) in GBM tissues were EDVs. They showed positive staining of CD34 (an endothelial marker) on the internal surface of the lumen and PAS on the vessel wall (Figure 1a and c), indicating the high angiogenic ability of GBM tissues. High microvascular proliferation was also observed by IF staining (Figure 1b and d). Abnormal nuclei were observed in some CD34-positive vessels (Figure 1b, indicated by white arrow).
EDVs in clinical and xenograft specimens of GBM. Representative images of EDVs in clinical (a) and xenograft (c) specimens detected using IHC staining. EDVs are both CD34- (brown) and PAS-positive (pink) on the vessel wall. Representative images of EDVs in clinical (b) and xenograft (d) specimens detected using IF staining. Endothelial cells are CD34-positive (green). GFAP (red) was used to identify GBM cells in clinical specimens and GSC-1 cells in xenograft specimens. DAPI (blue) stained the nucleus. The significant nuclear deformity was oobserved in CD34 positive cells in clinical specimens (b, indicated by white arrow). In contrast, the CD34 positve cells were normal at the cellular level in xenograft specimens (d, indicated by white arrow). Scale bar: 50 µm.
Abbreviations: EDV, Endothelium-dependent vessel; IHC, immunohistochemistry; IF, immunofluorescence; DAPI, 4′,6-diamidino-2-phenylindole; GBM, glioblastoma; GFAP, glial fibrillary acidic protein
ECMDVs appeared as PAS-positive vessels with no cell lining but with erythrocyte shadows in the lumen (Figure 2a and c). To verify the distribution of ECMDVs, we treated the tissue sections with an antibody against laminin (an extracellular matrix marker). As distinct from EDVs, the vessel wall of ECMDVs was stained positive for laminin but was negative with CD34 (Figure 2b and d).
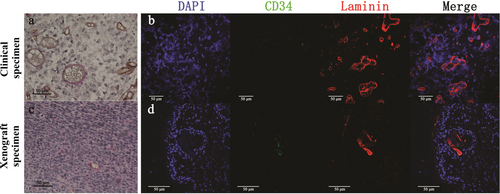
ECMDVs in clinical and xenograft specimens of GBM. Representative images of ECMDVs in clinical (a) and xenograft (c) specimens detected using IHC staining. ECMDVs are lined with extracellular matrix (marked by laminin; red) but not with endothelial cells (marked by CD34; brown). Red blood cells can be identified in the lumen of ECMDVs. Representative images of ECMDVs in clinical (b) and xenograft (d) specimens detected using IF staining. Endothelial cells are CD34-positive (green). Laminin (red) was used to identify extracellular matrix. DAPI (blue) stained the nucleus. Close loops of extracellular matrix are abundant in some area of the tissue sections. Some of them are connected with CD34 positive channels. Scale bar: 50 µm.
Abbreviations: ECMDV, extracellular matrix dependent vessel; IHC, immunohistochemistry; IF, immunofluorescence; DAPI, 4′,6-diamidino-2-phenylindole; GBM, glioblastoma
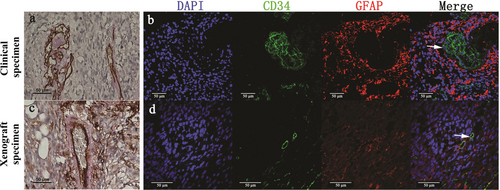
GDVs were also observed in tissue specimens containing red blood cells. They appeared to be lined by GBM cells (marked by PAS or GFAP) but not by endothelium (marked by CD34) when detected by IHC staining (Figure 3a and c). Similarly, GDVs were also observed in most of the tissue sections when detected by IF staining. As shown in Figure 3b and d, we observed GFAP-positive but CD34-negative staining of the cells surrounding the lumen in GDVs (expression of GFAP is a characteristic of GBM cells).
GDVs in clinical and xenograft specimens of GBM. Representative images of GDVs in clinical (a) and xenograft (c) specimens detected using IHC staining. GDVs appear as vessels lined by tumor-like cells with red blood cells in the lumen. These tumor-like cells are negative for CD34 (brown) and positive for PAS (pink). Representative images of GDVs in clinical (b) and xenograft (d) specimens detected using IF staining. The cells surrounding the GDV lumen are considered as tumor cells based on biomarkers and the morphology of their nuclei. GFAP (red) was used to identify GBM cells in clinical specimens as well as GSC-1 cells in xenograft specimens. The endothelial cells are CD34-positive (green). DAPI (blue) stained the nucleus. Scale bar: 50 µm.
Abbreviations: GBM, glioblastoma; GDV, GBM derived vessel; IHC, immunohistochemistry; IF, immunofluorescence; DAPI, 4′,6-diamidino-2-phenylindole
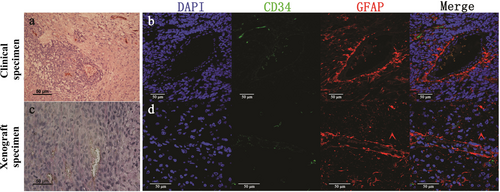
MVs are defined as blood vessels containing both GFAP-expressing tumor cells and CD34-positive endothelial cells in the same vessel wall. IHC staining of GBM specimens revealed the presence of MVs. Red cells were observed in some GDVs which were connected with EDVs in both clinical specimens and xenografts (Figure 4a and c). MVs contained endothelial cells (CD34-positive) and GFAP-positive GBM cells contributed to the formation of the vascular tubes in both human GBM specimens and mice xenografts (Figure 4b and d). The cells lined inside of GDVs were CD34 positive by IF staining, which were simillar to the GSC-1 cells expressing both CD34 and GFAP.
MVs in clinical and xenograft specimens of GBM. p>Representative images of MVs in clinical (a) and xenograft (c) specimens detected using IHC staining. MVs consist of CD34-negative and PAS-negative GBM derived vessels, as well as CD34-positive (brown) and PAS-positive (pink) endothelial vessels. Representative images of MVs in clinical (b) and xenograft (d) specimens detected using IF staining. One type of MVs is lined by GFAP-positive cells and CD34-positive cells. The other type is lined by GFAP and CD34 co-expressing cells. GFAP (red) was used to identify GBM cells in clinical specimens as well as GSC-1 cells in xenograft specimens. Endothelial cells are CD34-positive (green). DAPI (blue) stained the nucleus. Scale bar: 50 µm.
Abbreviations: GBM, glioblastoma; MV, mosaic vessel; IHC, immunohistochemistry; IF, immunofluorescence; DAPI, 4′,6-diamidino-2-phenylindole
3.2 MVD evaluation for four microcirculation patterns in GBM specimens
The MVDs determined by IHC and IF in both clinical and xenograft specimens are listed in Table 1. The MVD of four microcirculation patterns varied slightly and this depended on the detection method used. In the tissue sections stained by IHC, EDVs had the highest MVD and then followed by ECMDVs. However, in the tissue sections stained by IF, EDVs still had the highest MVD, and followed by MVs.
| Microvessel density (mean ± SD) | |||||
|---|---|---|---|---|---|
| Detection method | Sample type | EDV | ECMDV | GDV | MV |
| IHC | Clinical specimens | 7.88 ± 5.86 | 0.49 ± 1.09 | 0.20 ± 0.46 | 0.16 ± 0.40 |
| Xenograft specimens | 3.70 ± 2.20 | 0.50 ± 0.61 | 0.15 ± 0.22 | 0.40 ± 0.50 | |
| IF | Clinical specimens | 7.51 ± 5.55 | 0.25 ± 0.68 | 0.49 ± 0.80 | 0.76 ± 1.15 |
| Xenograft specimens | 3.55 ± 2.11 | 0.45 ± 0.60 | 0.05 ± 0.22 | 0.80 ± 0.70 | |
- Abbreviations: IHC, immunohistochemistry; IF, immunofluorescence; SD, standard error of the mean; EDV, endothelium-dependent vessel; ECMDV, extracellular matrix-dependent vessel; GDV, GBM cell-derived vessel; MV, mosaic vessel.
3.3 Three microcirculation patterns comprise a novel blood supply system in GBMs
All microcirculation patterns accessed in this study were observed in GBM specimens. Since both ECMDV and GDV are subtypes of VM [22], we analyzed VM vessels in GBM specimens by combining the data of ECMDV and GDV. The percentages of tissue samples that had VM were 38.8% (31/80) in clinical specimens and 20.0% (9/45) in xenograft specimens when detected by IHC. Additionally, when detected by IF, those percentages were 41.3% (33/80) and 20.0% (9/45), respectively.
To explore the 3D structures of the microcirculation patterns in GBM specimens, IF images of dual-stained sections were further analyzed by 3D reconstruction. GFAP-positive GBM cells lined the GDV walls (Figure 5a). Both CD34-negative and CD34-positive GBM cells, along with endothelial cells, lined the MV walls(Figure 5b and c). Moreover, CD34-positive endothelial cells formed EDVs (Figure 5a). GDVs connected with EDVs in some areas of the samples (Figure 5a). Our results therefore confirmed that MVs could be formed by GDVs and EDVs. Three microcirculation patterns (MVs, GDVs and EDVs) connected with each other and comprised a novel blood supply system for GBM.

Three-dimensional reconstruction of microcirculation patterns in GBM. a. The microcirculation patterns are not isolated from each other but inter-connect in three-dimensional structure. In a limited region of certain samples, only GDVs or EDVs can be observed. b-c. Two types of MVs can be identified clearly. One type is comprised of GFAP-positive tumor cells together with endothelial cells (b). The other type is comprised of GFAP and CD34 co-expressing tumor cells (c). Endothelial cells are CD34-positive (green). GFAP (red) was used to identify GBM cells. DAPI (blue) stained the nucleus.
Abbreviations: GBM, glioblastoma; GDV, GBM-derived vessel; EDV, endothelium-dependent vessel; MV, mosaic vessel; DAPI, 4′,6-diamidino-2-phenylindole
3.4 Patient characteristics and survival analysis
Sixty-four (80.0%) of the 80 GBM patients with complete follow-up data were selected for survival analysis. The median age of the patients was 47.6 years (range: 9.0-78.0 years), and 19 (29.7%) patients were female. The median Karnofsky Performance Status Score (KPS) was 78 (range: 20-90), and 15 (23.4%) patients had a KPS < 80 (Supplementary Table S2). All patients received standard radiotherapy and chemotherapy after surgery.
The methylation status of the MGMT promoter was detected in the samples from 61 of the 64 patients. We found that the MGMT promoter of 38 (59.4%) patients was methylated. IDH1/2 mutations were examined in eligible samples from 55 of the 64 patients, and 7 (10.9%) samples had IDH1 mutations, while none had IDH2 mutations.
The median survival of the 64 patients was 12.13 months (range: 1.20-149.50 months). Until the last follow-up, 3 (42.9%) of 7 patients with IDH1 mutations survived 149.50, 87.40, and 82.70 months, respectively. GBM patients with MGMT promoter methylation had a median survival of 12.33 months (range: 3.10-149.50 months) compared with 10.00 months (range: 1.20-82.70 months) for patients without MGMT promoter methylation (P = 0.037).
Patients with tumors containing GDVs had a median survival of 9.56 months (range: 1.20-22.83 months) compared with 13.60 months (range: 2.73-149.50 months) for patients without GDVs (P = 0.015; Figure 6a). In the MGMT promoter methylation cohort, patients with GDV-positive tumors had a median survival of 6.76 months (range: 3.53-20.93 months) compared with 14.23 months (range: 3.13-149.50 months) for patients with GDV-negative tumors (P = 0.022; Figure 6b). In the IDH1 wild-type cohort, patients with GDV-positive tumors had a median survival of 10.00 months (range: 1.20-20.93 months) compared with 13.60 months (range: 2.73-83.20 months) for patients with GDV-negative tumors (P = 0.096; Figure 6c). We performed multivariable analysis including gender, age and MGMT promoter methylation status for the prognosis of GDVs. The results showed that the presence of GDVs predicted a poor prognosis for GBM patients (hazard ratio [HR] = 2.21; 95% confidence interval [CI] = 1.13-4.32; P = 0.020), especially for the patients with MGMT promoter methylation (HR = 1.178; 95% CI = 0.65-2.11; P = 0.018). The results of survival analysis for GBM patients with other microcirculation patterns in tumors are summarized in Supplementary Table S3. GDVs might be a negative predictor for the survival of GBM patients, even in those with MGMT promoter methylation.
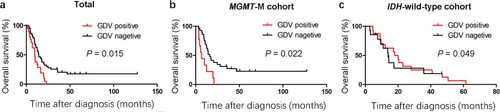
Kaplan-Meier survival curves of the GBM patients. a. Patients with tumors containing GDVs had a median survival of 9.56 months (range: 1.20-22.83 months) compared with 13.60 months (range: 2.73-149.50 months) for patients with tumors without GDVs (P = 0.015). b. In the MGMT promoter methylation cohort, patients with GDV-positive tumors had a median survival of 6.76 months (range: 3.53-20.93 months) compared with 14.23 months (range: 3.13-149.50 months) for patients with GDV-negative tumors (P = 0.022). c. In the IDH1 wild-type cohort, patients with GDV-positive tumors had a median survival of 10.00 months (range: 1.20-20.93 months) compared with 13.60 months (range: 2.73-83.20 months) for patients with GDV-negative tumors (P = 0.096)
4 DISCUSSION
In a previous study, three microcirculation patterns, VMs, MVs, and EDVs, were identified in gliomas [23]. In the present study, four microcirculation patterns, EDVs, ECMDVs, GDVs, and MVs, were observed in both clinical GBM and xenograft specimens. GDV and ECMDV are subtypes of VM vessels. The present study confirmed that EDVs were the dominant blood vessels in all GBM specimens, constituting more than 80% of blood vessels in GBM tissues. During tumor growth, hypoxia may induce VEGF expression to stimulate endothelial proliferation, leading to angiogenesis in malignant tumors [24]. In the present study, three other microcirculation patterns (ECMDVs, GDVs, and MVs) were observed in some but not all GBM specimens, indicating that these vessels might compensate for the GBM microcirculation. Although the constituent ratios of each microcirculation pattern in each sample varied, the sum of the three patterns was about 10%, while the EDV ratio was greater than 80%. This finding was confirmed by our data obtained from both IF and IHC stainings.
Our findings hint that compensatory microcirculation (ECMDVs, GDVs, and MVs) might play important roles when considering the failure of bevacizumab in treating GBM. Even if bevacizumab inhibits conventional angiogenesis by targeting EDVs, tumor cells can still acquire enough blood supply via these complementary microcirculation vessels. Among these complementary microcirculation vessels, GDVs may be the most critical component with the potential to initiate a new blood supply system in GBM.
Microcirculation patterns were more easily revealed in GBM by using IF staining compared to IHC staining. IF staining should be an appropriate method to identify GDVs at the cell level [25] since some GBM cells may express CD34 and contribute to blood vessel formation. The differentiation ability of GBM stem-like cells is not surprising and is a possible way of angiogenesis in GBMs [18]. CD34 serves as a biomarker of endothelial progenitor cells [26]. We previously observed CD34-positive GDECs and GDEC vessels in glioma tissue samples [18]. We also detected CD34- and GFAP-coexpressing cells in the vessel wall. CD34-positive cells are not exclusively endothelial cells because the nuclei of some CD34-positive cells are enlarged and abnormal [27]. We also examined the varying capacity of GBM cells to differentiate in vitro and recapitulate the network patterns observed in tissue sections in the absence of endothelial cells in a previous study [28]. CD34 is expressed in stem cells [29, 30]. Using live-cell imaging, we observed CD34 expression in some GBM stem-like cells when they were treated with VEGF. CD34-positive GBM cells that form a special type of VM channel may represent a subset of malignant GBM cells. EDVs should contain GDEC vessels, because GDECs may completely differentiate into endothelial cells, thus, MV may represent an intermediate status between GDVs and EDVs.
We also propose an independent association between GDVs and survival of GBM patients. Our data indicated that MGMT promoter methylation was independently associated with a favorable outcome in patients with GBM, which is consistent with previous study [31]. However, in MGMT promoter methylation cohort, patients with GDV-positive tumors had a significantly shorter survival than patients without GDVs, suggesting that GDVs may contribute to the poor prognosis of patients with GBM. This finding may partially explain the failure of anti-angiogenic therapy with Avastin. Even if EDV generation is blocked by targeted therapy [32], GDV generation can maintain the blood supply which is needed for tumor growth. We found that ECMDVs and MVs were not associated with the prognosis of patients with GBM, and the formation of GDV channels should be the key process in the formation of a new blood supply network during GBM growth [14].
Our future work will keep deciphering the mechanism of GDV formation and figuring out the potential anti-angiogenic targets in GBM. As a supplementary treatment strategy, anti-angiogenic therapy targeting the markers of VM might represent a novel choice to suppress the blood supply and subsequently block the proliferation of GBM. Furthermore, constricting the dynamic profile of microcirculation patterns in the evolution of GBM is of importance.
5 CONCLUSIONS
The present study observed four microcirculation patterns, EDV, ECMDV, GDV, and MV, in GBM specimens and found that GDVs might serve as a negative predictor for survival of GBM patients, even in the patients with MGMT promoter methylation. GDVs may play an important role in forming a new blood supply system for GBM tissues. Therefore, targeting GDVs might provide a novel strategy to block blood supply in GBM.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
All procedures performed in this study involving animal experiments were in accordance with the ethical standards of the Accreditation of Laboratory Animal Care and approved by the Ethics Committee of Sun Yat-sen University Cancer Center (GZR2013-052).
CONSENT FOR PUBLICATION
Not applicable.
COMPETING INTERESTS
The authors declare that they have no competing interests.
FUNDING
This study was supported by the grants from the National Basic Research Program of China (No. 2015CB755505), the National Natural Science Foundation of China (No. 30973478, 81372685, 81572479, and 81672484), the Guangzhou Science Technology Project (No. 201508020125 and 201803010056), the Science and Technology Planning Project of Guangdong Province (No. 2016A020213004), the Natural Science Foundation of Guangdong Province (No. S2013040012894), and the Shenzhen Innovation Project of Scientific and Technology (No. JCYJ20140416094330210).
AUTHORS’ CONTRIBUTIONS
ZPC: Planned, wrote, revised, and supervised the writing and concept of the article. XM wrote and edited the manuscript, constructed xenograft animal model and analyzed the GBM samples. JW analyzed the data. YSC, QPZ, FRC, JZ, and HPC reviewed and edited the manuscript. SYX and YKL reviewed and edited the manuscript, performed IHC and IF staining, identified IDH mutation and MGMT methylation state. CK reviewed and edited the manuscript with comments. All authors read and approved the final manuscript.
ACKNOWLEDGEMENTS
We sincerely appreciate the generous help from the core facility in the Department of Experimental Research, Sun Yat-sen University Cancer Center.
Open Research
AVAILABILITY OF DATA AND MATERIALS
The datasets used and/or analyzed in the current study are available from the corresponding author on reasonable request or through Research Data Deposit (http://www.researchdata.org.cn), the approval number is RDDB2019000750.




