Emerging role of deep learning-based artificial intelligence in tumor pathology
Abstract
The development of digital pathology and progression of state-of-the-art algorithms for computer vision have led to increasing interest in the use of artificial intelligence (AI), especially deep learning (DL)-based AI, in tumor pathology. The DL-based algorithms have been developed to conduct all kinds of work involved in tumor pathology, including tumor diagnosis, subtyping, grading, staging, and prognostic prediction, as well as the identification of pathological features, biomarkers and genetic changes. The applications of AI in pathology not only contribute to improve diagnostic accuracy and objectivity but also reduce the workload of pathologists and subsequently enable them to spend additional time on high-level decision-making tasks. In addition, AI is useful for pathologists to meet the requirements of precision oncology. However, there are still some challenges relating to the implementation of AI, including the issues of algorithm validation and interpretability, computing systems, the unbelieving attitude of pathologists, clinicians and patients, as well as regulators and reimbursements. Herein, we present an overview on how AI-based approaches could be integrated into the workflow of pathologists and discuss the challenges and perspectives of the implementation of AI in tumor pathology.
Abbreviations
-
- AI
-
- artificial intelligence
-
- AR
-
- androgen receptor
-
- ATC
-
- anaplastic thyroid carcinoma
-
- AUC
-
- area under receiver operating characteristic curve
-
- CLIA
-
- Clinical Laboratory Improvement Amendment
-
- CNN
-
- convolutional neural network
-
- CTC
-
- circulating tumor cell
-
- DL
-
- deep learning
-
- DSS
-
- disease-specific survival
-
- EGFR
-
- epidermal growth factor receptor
-
- ER
-
- estrogen receptor
-
- FAT1
-
- FAT atypical cadherin 1
-
- FCN
-
- fully convolutional network
-
- FDA
-
- Food and Drug Administration
-
- FTC
-
- follicular thyroid carcinoma
-
- GAN
-
- generative adversarial network
-
- HE
-
- hematoxylin and eosin
-
- HER2
-
- human epidermal growth factor receptor 2
-
- HGUC
-
- high-grade urothelial carcinoma
-
- HP
-
- hyperplastic polyp
-
- HPF
-
- high power field
-
- HR
-
- hazard ratio
-
- KRAS
-
- Ki-ras2 Kirsten rat sarcoma viral oncogene homolog
-
- ML
-
- machine learning
-
- MSI
-
- microsatellite instability
-
- MSS
-
- microsatellite stability
-
- MTC
-
- medullary thyroid carcinoma
-
- OS
-
- overall survival
-
- PD-L1
-
- programmed death-ligand 1
-
- PTC
-
- papillary thyroid carcinoma
-
- RNN
-
- recurrent neural network
-
- ROI
-
- region of interest
-
- SETBP1
-
- SET binding protein 1
-
- SPOP
-
- speckle-type POZ protein
-
- SSAP
-
- sessile serrated adenoma/polyp
-
- STK11
-
- serine/threonine kinase 11
-
- TCGA
-
- The Cancer Genome Atlas
-
- TSA
-
- traditional serrated adenoma
-
- WSI
-
- whole-slide image
1 BACKGROUND
Artificial intelligence (AI) was termed by McCarthy et al. [1] in the 1950s, referring to the branch of computer science in which machine-based approaches were used to make predictions to mimic what human intelligence might do in the same situation. AI, currently a hot and controversial topic, has been introduced into many aspects of our everyday life, including medicine. Compared with other applications in the treatment of diseases, AI is more likely to enter the diagnostic disciplines based on image analysis such as pathology, ultrasound, radiology, and ophthalmic and skin disease diagnosis [2, 3]. Among these applications, the implementation of AI in pathology presents a special challenge due to the complexity and great responsibility of pathological diagnosis.
The progress of AI in pathology depended on the growth of digital pathology. In the 1960s, Prewitt et al. [4] scanned simple images from a microscopic field of a common blood smear and converted the optical data into a matrix of optical density values for computerized image analysis, which is regarded as the beginning of digital pathology. After the introduction of whole-slide scanners in 1999, AI in digital pathology using computational approaches grew rapidly to analyze the digitized whole-slide images (WSIs). The creation of large-scale digital-slide libraries, such as The Cancer Genome Atlas (TCGA), enabled researchers to freely access richly curated and annotated datasets of pathology images linked with clinical outcome and genomic information, in turn promoting the substantial investigations of AI for digital pathology and oncology [5, 6]. Our group identified an integrated molecular and morphologic signature associated with chemotherapy response in serous ovarian carcinoma using TCGA data in 2012 [7], which contains rudimentary model of machine learning (ML) on WSIs of TCGA.
AI models in pathology have developed from expert systems to traditional ML and then to deep learning (DL). Expert systems rely on rules defined by experts, and traditional ML needs to define features based on expert experience, while DL directly learns from raw data and leverages an output layer with multiple hidden layers (Figure 1) [8]. Compared with expert systems and hand-crafted ML approaches, DL approaches are easier to be conducted and have high accuracy. The increase in computational processing power and blooming of algorithms, such as convolutional neural network (CNN), fully convolutional network (FCN), recurrent neural network (RNN), and generative adversarial network (GAN), have led to multiple investigations on the usage of DL-based AI in pathology. The application of AI in pathology helps to overcome the limitations of subjective visual assessment from pathologists and integrate multiple measurements for precision tumor treatment [9].
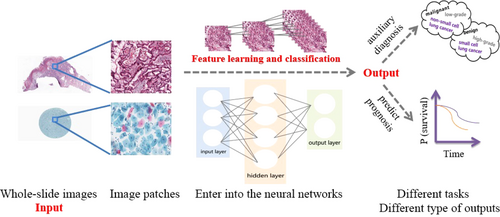
The Food and Drug Administration (FDA) in the USA approved the Philips IntelliSite whole-slide scanner (Philips Electronics, Amsterdam, Netherlands) in 2017, which is a milestone for true digital pathology laboratories. The application of AI in pathology is also promoted by startups, for example, PAIGE.AI [10], Proscia [1], DeepLens [12], PathAI [13] and Inspirata [14], who are using DL-based AI tools for detecting, diagnosing and predicting the prognosis of several types of cancers. Several institutions have decided to digitize their entire pathology workflow [15-17]. Therefore, the application of AI in pathology has been on the road. In this review, we summarized current studies on DL-based AI application in tumor pathology, analyzed the advantages and challenges of AI application, and discussed the optimized workflow and perspectives of human-machine cooperation in tumor pathology.
2 APPLICATION OF DL-BASED AI IN TUMOR PATHOLOGY
The implementation of AI in tumor pathology refers to almost all kinds of tumors (Table 1) and is involved in tumor diagnosis, subtyping, grading, staging, and prognosis prediction as well as the identification of pathological features, biomarkers, and genetic changes.
| Applications | The diagnostic points of AI in the studies | Performance | References |
|---|---|---|---|
| Breast cancer | |||
| Diagnosis | Carcinoma/Non-carcinoma | Accuracy: 83.3% | [18] |
| Normal/Benign/Carcinoma in situ/Invasive carcinoma | Accuracy: 77.8% | [18] | |
| Non-malignant/Malignant | AUC: 0.962 | [19] | |
| Benign/Ductal carcinoma in situ/Invasive ductal carcinoma | Accuracy: 81.3% | [19] | |
| Invasive cancer/Benign lesion based on both tumor cells and stroma | AUC: 0.962 | [20] | |
| Non-proliferative/Proliferative/Atypical hyperplasia/Carcinoma in situ/Invasive carcinoma | Precision: 81% | [24] | |
| Tumor subtyping | Adenosis/Fibroadenoma/Tubular adenoma/Phyllodes tumor/Ductal carcinoma/Lobular carcinoma/Mucinous carcinoma/Papillary carcinoma | Accuracy: 90.66%-93.81% | [37] |
| Tumor grading | Low/Intermediate/High grade | Accuracy: 69% | [42] |
| Tumor staging | Heatmap for the region of invasive cancer | Dice coefficient: 75.86% and 76.00% | [44], [45] |
| With/Without lymph node metastasis | AUC: 0.994 and 0.996 | [46], [47] | |
| Metastatic regions in lymph node | Sensitivity: 0.807-0.910 | [46]-[48] | |
| Evaluation of pathological features | Mitotic count | F-score: 0.611 | [57] |
| Ki-67 index | F-score: 0.91 | [64] | |
| Proliferation score | Kappa: 0.613 | [58] | |
| Immune cell-rich/Immune cell-poor regions | AUC: 0.99 | [62] | |
| Evaluation of biomarkers | HER2 status: Negative (0 and 1+)/ Equivocal (2+)/Positive (3+) | Accuracy: 83% and 87% | [66], [67] |
| Lung cancer | |||
| Tumor subtyping | Small cell cancer/Non-small cell cancer (Cytology) | Accuracy: 85.6% | [39] |
| Adenocarcinomas/Squamous cell carcinomas/Small cell carcinomas (Cytology) | Accuracy: 60%-89% | [39] | |
| Adenocarcinoma/Squamous cell carcinoma | AUC: 0.83-0.97 | [31] | |
| Solid/Micropapillary/Acinar/Cribriform subtype | F-score: 0.60-0.96 | [32] | |
| Lepidic/Solid/Micropapillary/Acinar/Cribriform subtype | AUC: 0.961-0.997 | [33] | |
| Evaluation of pathological features | Immune cell count | Accuracy: 98.6% | [63] |
| Evaluation of biomarkers | PD-L1 status: Negative/Positive | AUC: 0.80 | [69] |
| Evaluation of genetic changes | Predict the most commonly mutated genes | AUC: 0.733-0.856 | [31] |
| Prognosis prediction | Low/High risk | Hazard ratio: 2.25 | [76] |
| Colorectal cancer | |||
| Diagnosis | Benign/Malignant | Accuracy: 95%-98% | [21] |
| Normal/Cancer | Accuracy: 97% | [22] | |
| Tumor subtyping | HP/SSAP/TSA/Tubular adenoma/Tubulovillous and villous adenoma | Accuracy: 93% | [34] |
| Tumor grading | Normal /Low-grade cancer/High-grade cancer | Accuracy: 91% | [22] |
| Evaluation of pathological features | Number of tumor budding | Correlation R2: 0.86 | [59] |
| Evaluation of genetic changes | MSI/MSS on HE stained images | AUC: 0.77-0.84 | [73] |
| Prognosis prediction | DSS, OS: Low/High risk | Hazard ratio: 1.65-2.30 | [73]-[75] |
| Gastric cancer | |||
| Diagnosis | Normal/Dysplasia/Cancer | Accuracy: 86.5% | [25] |
| Evaluation of genetic changes | HER2 status: Negative (0 and 1+)/Positive (2+ and 3+) | Accuracy: 69.9% | [68] |
| Prostate cancer | |||
| Tumor grading | Gleason scoring | Accuracy: 75% | [40] |
| Evaluation of genetic changes | Distinguish SPOP mutant from non-mutant on HE stained images | AUC: 0.74-0.86 | [70] |
| Cervical cancer | |||
| Diagnosis | Normal/Abnormal (Cytology: smear-based and liquid-based) | Accuracy: 98.3% and 98.6% | [27] |
| Tumor subtyping | Keratinizing/Non-keratinizing/Basaloid squamous cell carcinoma | Accuracy: 93.33% | [38] |
| Glioma | |||
| Tumor grading | Grade IV/Grade II/III (Glioblastoma multiform) | Accuracy: 96% | [41] |
| Grade II/Grade III | Accuracy: 71% | [41] | |
| Prognosis prediction | OS: Low/Intermediate/High risk | Concordance index: 0.754 | [77] |
| Thyroid cancer | |||
| Diagnosis | PTC/Benign nodules (Cytology) | Accuracy: 97.66% | [30] |
| Tumor subtyping | Normal tissue/Adenoma/Nodular goiter/PTC/FTC/MTC/ATC | Accuracy: 88.33%-100% | [36] |
| Others | |||
| Diagnosis for esophagus lesion | Barrett esophagus/Dysplasia/Cancer | Accuracy: 83% | [26] |
| Diagnosis for melanocytic lesion | Nevus/aggressive malignant melanoma | AUC: 0.998 | [23] |
| Diagnosis for urinary tract lesion | HGUC and Suspicious HGUC/other lesions (Cytology) | AUC: 0.88 and 0.92 | [28], [29] |
| Subtyping for ovary cancer | Serous/Mucinous/Endometrioid/Clear cell carcinoma | Accuracy: 78.2% | [35] |
| Staging for osteosarcoma | Regions of tumor component/Necrosis/non-tumor component | Accuracy: 92.4% | [43] |
| Biomarker for pancreatic neuroendocrine neoplasm | Segment tumor regions to calculate Ki-67 index | Accuracy: 96.2% | [65] |
| Multiple tasks in breast cancer | ER status:Negative/Positive | Accuracy: 84% | [78] |
| RNA-based molecular subtypes: Basal-like/Non-basal-like | Accuracy: 77% | ||
| Histological subtypes: Ductal/Lobular | Accuracy: 94% | ||
| Grade: Low-intermediate grade/High grade | Accuracy: 82% | ||
| Recurrence-risk: High/Low-medium risk | Accuracy: 76% | ||
| Multiple cancers | |||
| Lung/Breast/Bladder cancer | Different cancer types: Lung/Breast/Bladder cancer | Accuracy: 100% | [79] |
| Adenocarcinoma/Squamous cell carcinoma of lung cancer | Accuracy: 92% | ||
| Identifying biomarkers based on expression pattern | Accuracy: 95% | ||
| Scoring biomarkers | Accuracy: 69% | ||
| Prostate/Lung cancer | CTC status: Positive/Negative | Accuracy: 84% | [56] |
- Abbreviations: AUC, area under receiver operating characteristic curve; HE, hematoxylin and eosin; HP, hyperplastic polyp; SSAP, sessile serrated adenoma/polyp; TSA, traditional serrated adenoma; MSI/MSS, microsatellite instability/stability; DSS, disease-specific survival; OS, overall survival; PTC/FTC/MTC/ATC, papillary/follicular/medullary/anaplastic thyroid carcinoma; HGUC, high-grade urothelial carcinoma; ER, estrogen receptor; CTC: circulating tumor cell.
2.1 Tumor diagnosis
It is the most important for pathologists to distinguish tumors from other lesions and distinguish malignant from benign tumors as these can directly affect treatment decisions for different therapeutic strategies. Araújo et al. [18] developed a CNN-based AI algorithm to classify breast WSI images into two categories (carcinoma and non-carcinoma) with an accuracy of 83.3% and into four categories (normal tissue, benign lesions, carcinoma in situ, and invasive carcinoma) with an accuracy of 77.8%. Bejnordi et al. [19] developed a context-aware stacked CNN which was first trained to recognize the lower-level features and then used as input data to train the next stacked network to recognize the higher-level features. Using this CNN, they obtained an area under the receiver operating characteristic curve (AUC) of 0.962 to distinguish breast malignant tumors from non-malignant lesions and an accuracy of 81.3% to classify benign lesions, ductal carcinoma in situ, and invasive ductal carcinoma. Considering the effect of stroma on tumors, Ehteshami Bejnordi et al. [20] designed a CNN-based model to combine stroma features to distinguish breast invasive cancer from benign lesions. DL-based AI algorithms also gained comparable accuracy with professional pathologists in discriminating between benign and malignant colorectal tumors [21, 22], and melanoma from nevus [23].
Using the weakly supervised DL models that dramatically reduced labeling workload, Mercan et al. [24] classified breast lesions into non-proliferative, proliferative, atypical hyperplasia, carcinoma in situ, and invasive carcinoma using WSIs of breast biopsy specimens with a precision of 81%. Wang et al. [25] classified gastric lesions into normal, dysplasia, and cancer with an accuracy of 86.5% and Tomita et al. [26] classified esophagus lesions into Barrett esophagus, dysplasia, and cancer with an accuracy of 83%.
In addition to biopsy and resection specimens, pathologists should perform cytology diagnosis in routine work. For cervical cytological diagnosis, AI could classify cells as normal or abnormal in smear-based and liquid-based images, reaching an accuracy of 98.3% and 98.6%, respectively [27]. In liquid-based urine cytology, AI could separate high-grade urothelial carcinoma (HGUC) and suspicious HGUC from other lesions based on cell level-features [28] or WSI level-features [29]. AI also performed showed promising ability in the differential diagnosis for thyroid tumors on the basis of cytological images [30]. In summary, DL-based AI has shown promise in histological and cytology diagnosis for many kinds of tumors.
2.2 Tumor subtyping
The therapeutic strategies are different in various subtypes of cancers. A CNN-based model automatically differentiated lung adenocarcinoma, squamous cell carcinoma and normal lung tissue on images from biopsies, frozen tissues and formalin-fixed paraffin-embedded tissues with a high AUC (0.83-0.97) [31]. Considering different growth patterns of invasive lung adenocarcinoma cells are related with the clinical outcomes of patients, Gertych et al. [32] and Wei et al. [33] developed CNN algorithms to sort each image tile into individual growth pattern and generate a probability map for a WSI, facilitating pathologists to quantitatively report the major and more malignant components of lung adenocarcinoma, such as micropapillary and solid components. Similarly, DL-based AL were performed for multi-categorization of colorectal polyp [34], ovarian cancer [35], thyroid tumor [36], breast tumor [37], and cervical squamous cell carcinoma [38]. On the basis of cytological image, AI could recognize the histological subtypes of lung cancer with an accuracy of 60%-89% [39].
2.3 Tumor grading
Pathologists often evaluate tumor grades according to the differentiation of tumor cells, glandular architecture, mitosis, necrosis and more, which are subjective and may possibly skew treatment decision and clinical monitoring. Arvaniti et al. [40] proposed an AI model for the Gleason scoring of prostate cancer; suitable for any Gleason scores. The agreement between the AI model and each of the two pathologists (0.75 and 0.71, respectively) was comparable to that between the two pathologists themselves (0.71). Moreover, the AI model was superior to the pathologists in distinguishing low-risk from intermediate-risk cases based on the Gleason scores calculated by the model. Ertosun et al. [41] developed two separate CNNs to define the grade of gliomas. One of them classified the cases as either glioblastoma multiform (grade IV) or lower grade glioma (grade II and III), with an accuracy of 96%, and the other discriminated grade II glioma from grade III with an accuracy of 71%. For the breast biopsy images, a CNN-based model distinguished low-, intermediate-, and high-grade breast cancers with an accuracy of 69% [42]. DL-based models were also set up to discriminate normal tissue, low-grade and high-grade colorectal adenocarcinoma with an accuracy of 91% [22]. Overall, AI will be helpful to provide objective and reproducible results for tumor grading.
2.4 Tumor staging
For resection samples, pathologists should provide as much information possible for TNM staging that usually determine therapeutic decisions. In osteosarcoma, a CNN-based model distinguished three types of region of interests (ROIs), namely the tumor, necrotic and non-tumor component (e.g., bone, cartilage), on a patch level (64,000 patches from 82 WSIs) with an accuracy of 92.4% [43]. In addition, the proportion of necrosis, a variable factor for prognosis, could be calculated. Several DL-based networks have also been developed to recognize tumor regions in breast cancer [24, 44, 45].
The assessment of lymph node metastasis is necessary for tumor staging but it is time-consuming and error-prone for pathologists. In the “Cancer Metastases in Lymph Nodes Challenge” (CAMELYON16), a competition between AI and pathologists to assess sentinel lymph nodes of breast cancer, two AI algorithms exceeded pathologists with the best AUC of 0.994 in slide-level detection (only identifying metastasis or not), and two algorithms outperformed pathologists with the best one achieving the mean sensitivity across six false-positive rates of 0.807 in lesion-level detection (recognizing all metastases except for isolated tumor cells) [46]. In the same dataset, Lymph Node Assistant (LYNA), a more optimized algorithm, gained a higher AUC (0.996) in slide-level detection and a higher sensitivity with one false positive per slide (91%) in lesion-level detection through filtering out artifacts. Of note, LYNA corrected two slides misdiagnosed as “normal” by the organizers [47]. Another study demonstrated that LYNA improved the sensitivity of detecting all micrometastases in lymph nodes from 83% to 91% (P = 0.02) with a significantly shorter review time as compared to pathologists alone [48].
In the past decade, numerous studies have shown that circulating tumor cells (CTCs) may be used as a marker to predict disease progression and survival in metastatic [49-51] and possibly even in early-stage cancer patients [52]. High CTC numbers were correlated with aggressive disease, increased metastasis, and decreased time to relapse [53]. Considering the simplicity and minimally invasion of blood collection, CTCs are expected to be used as a marker for monitoring tumor progression and guiding therapeutic management as well as indicating therapy effectiveness [54]. However, technical obstacles, such as small quantity and lack of standardized detection assays and valid marker limit clinical usage [55]. Zeune et al. [56] reported that the manual counting for CTCs of prostate cancer and non-small cell lung cancer on fluorescent images varied among human reviewers and counting platforms, whereas DL-based CTC recognition was relatively stable with higher accuracy than the average level of human reviewers.
Considering the current contribution of AI to identifying tumor regions, detecting lymph node metastasis and CTCs, and the ability of AI to analyze large amounts of data, AI approaches will potentially help pathologists and oncologists perform tumor staging.
2.5 Evaluation of pathological features
Mitosis represents the proliferation ability of tumor cells. However, mitosis count is time-consuming. The Assessment of Mitosis Detection Algorithms 2013 (AMIDA13) generated a good algorithm recognizing mitoses with the F1-score of 0.611 on 1000 images at high power fields (HPFs) from breast cancers, which was comparable to the performance of inter-observers [57]. The proliferation scores of breast cancers were reported based on the WSIs-level AI detection in the Tumor Proliferation Assessment Challenge 2016 (TUPAC16) [58].
Tumor budding is one of the aggressive behavior of tumors. In colorectal carcinomas, Weis et al. [59] utilized CNN to obtain the absolute number of tumor budding based on cytokeratin-stained WSIs and demonstrated the correlation between the number of budding hotspots and the status of lymph node. It has been reported that the kind and amount of tumor-infiltrating immune cells are related with the sensitivity to immunotherapy and prognostic stratification for tumor patients [60, 61]. A DL method using CD45-annotated digital images could quantify immune cells and distinguish immune cell-rich or -poor regions in breast cancer [62]. The AUC was 0.99 without the limitation of histological types and grades. The AI algorithm had an excellent performance in counting T-cells and B-cells on both cell patch- and WSI-level images of CD3-, CD8-, and CD20-stained sections from lung tissue, undisturbed by anthracitic pigment [63]. Compared with the hand-crafted AI focused on hand-crafted pathological features, DL-based AI could recognized both hand-crafted features and domain-agnostic features which could be applied across disease and tissue types.
2.6 Evaluation of biomarkers
Saha et al. [64] used a DL-based approach to automatically detect high-proliferation regions and calculate the Ki-67 index of breast cancer. Niazi et al. [65] developed AI algorithms to segment tumor areas from interstitium and normal pancreatic tissue on the unevenly Ki-67-immunoreactive WSIs and accordingly calculated the Ki-67 index more precisely in pancreatic neuroendocrine neoplasms.
Furthermore, some biomarkers are used for the selection of suitable patients for the related therapies. Trastuzumab could be used on the basis of the status of human epidermal growth factor receptor 2 (HER2) in breast and gastric cancer. In breast cancer, a CNN-based algorithm achieved an overall agreement of 83% with a pathologist which was similar to the agreement among pathologists in predicting HER2 negative (0 and 1+), equivocal (2+), and positive (3+) on WSIs [66]. The prediction results of AI improved when investigators segmented cell membranes as the true expression location of HER2 [67]. Similarly, AI algorithm was also developed to assess HER2-positive areas (2+ and 3+), HER2-negative areas (0 and 1+) and tumor-free areas in gastric cancer with an accuracy of 69.9% [68]. To select potential patients sensitive to pembrolizumab, an AI algorithm identified the expression of programmed death-ligand 1 (PD-L1; negative or positive) on hematoxylin and eosin (HE) stained images of non-small cell lung cancer with AUC of 0.80 which was comparable to the assessment of pathologists based on PD-L1 immunohistochemistry images [69]. Based on the immunohistochemical staining or fluorescent staining WSIs, even on HE staining WSIs, DL-based AI algorithms performed the evaluation of biomarkers, which were involved in diagnosis, prognosis and drug response prediction.
2.7 Evaluation of genetic changes
The morphological changes shown in WSI are manifestations of underlying genetic changes. To predict whether or not the speckle-type POZ protein (SPOP) gene is mutated in prostate cancer, Schaumberg et al. [70] trained multiple ensembles of residual networks using a cohort of 177 prostate cancer patients from TCGA where 20 had mutant SPOP and validated their findings in an independent cohort from MSK-IMPACT of 152 patients where 19 had mutant SPOP. Despite the training set was from frozen sections and the validation set was from formalin-fixed paraffin-embedded sections, mutants and non-mutants of SPOP were accurately distinguished (AUC = 0.86). Considering that non-mutant SPOP ubiquitinylates androgen receptor (AR) to mark AR for degradation [71], identification of SPOP mutation state could lead directly to precision medicine. Moreover, SPOP mutation is mutually exclusive with TMPRSS2-ERG gene fusion [72], the prediction of SPOP mutation status provided indirect information of the TMPRSS2-ERG state and potentially others. Coudray et al. [31] trained the DL network to predict the ten most commonly mutated genes in lung adenocarcinoma from TCGA pathology images and found that six of them (serine/threonine kinase 11 [STK11], epidermal growth factor receptor [EGFR], FAT atypical cadherin 1 [FAT1], SET binding protein 1 [SETBP1], Ki-ras2 Kirsten rat sarcoma viral oncogene homolog [KRAS] and TP53) could be predicted with AUCs from 0.733 to 0.856. Additionally, an AI model was developed to identify microsatellite instability (MSI) or microsatellite stability (MSS) based on HE stained images of gastrointestinal cancer without performing microsatellite instability assays, which also showed robustness in snap-frozen samples, Asian populations, and even endometrial cancer with high AUC (0.77-0.84) [73]. Through these AI networks, patients with specific genetic changes were identified based on intrinsic genetic-histologic relationships, which benefited the precision treatment.
2.8 Prognosis prediction
Bychkov et al. [74] used HE-stained tissue microarray images of colorectal cancer to invent a DL-based method to divide the patients into low- or high-risk groups. It is noticeable that the annotation in the training set was patients’ outcome rather than the labeled WSIs. The AI exceeded histological grade (hazard ratio [HR], 2.30 vs. 1.65) and was demonstrated to be an independent prognostic factor with multivariate Cox proportional hazard model analysis. Kather et al. [75] showed that integrated interstitium characteristics (including adipose, debris, lymphocytes, muscle, and desmoplastic stroma) extracted by CNN could independently forecast the overall survival (HR, 2.29) and relapse-free survival (HR, 1.92) of patients with colorectal cancer in multi-centers dataset, regardless of clinical stage. It was demonstrated that DL approaches could predict the prognostic risk by learning histologic features in lung adenocarcinoma [76] and glioma [77]. Kather et al. [73] reported that their algorithm-based prediction of MSI was also fit for predicting overall survival in gastrointestinal cancer. The above investigations indicated that AI algorithms could be used to predict clinical outcome besides pathological diagnosis for cancer patients.
2.9 Algorithm for multiple tasks and multiple tumors
Most of the above AI algorithms were designed to conduct a specific job in a certain tumor type. As a result, the pathologists need to run many AI algorithms to complete a pathological report, in which the tumor were to be diagnosed, subtyped, graded by different algorithms and the high-risk features were to be assessed by another individual algorithms.
Couture et al. [78] developed a DL-based algorithm to complete multiple tasks in HE-stained tissue microarray images of breast cancer, including identifying histological subtypes (ductal or lobular) with an accuracy of 94%, classifying histological grade (low-intermediate grade or high grade) with an accuracy of 82%, and assessing the status of estrogen receptor (ER; positive or negative) with an accuracy of 84%, discriminating molecular subtypes (basal-like or non-basal-like types) with an accuracy of 77%, as well as stratifying recurrence-risk (high or low-to-medium) with an accuracy of 76%. Another system equipped with six state-of-the-art DL architectures has the potential to determine tumor, subtypes, as well as biomarkers’ score in lung, breast, and bladder cancer [79].
3 CHALLENGES AND PERSPECTIVES
As displayed in the above explorations, DL-based AI has appealing perspective for elevating the efficiency of pathological diagnosis and prognosis. However, there are still some obstacles and challenges in the implementation of AI in tumor pathology.
3.1 Validation
Current AI algorithms are mainly established on small-scale data and images from single-center. The data from single center were still deviation, although researchers have developed methods to augment the dataset, including but not limited to random rotation and flipping, color jittering, and Gaussian blur [32, 35, 38, 39]. Variations exist in slide preparation, scanner models and digitization among different centers. Zech et al. [80] reported that a CNN for pneumonia detection performed significantly poorer when it was trained using data from one institution and validated independently using data from two other institutions than when it was trained using data from all three institutions (P < 0.001). AI algorithms need to be sufficiently validated using multi-institutional data before clinical adoption.
Fortunately, we can find some well-curated, accurate WSI reference datasets across cancer subtypes with annotated cancerous regions, which contribute to standardizing the evaluation of AI algorithms. Moreover, we can use some large-scale digital-slide libraries, including TCGA, like training or validation datasets. Building comprehensive quality control and standardization tools, data share and validation with multi-institutional data can increase the generalizability and robustness of the AI algorithms. In addition, AI algorithms need to be continually validated and corrected by the diagnosis of expert pathologists.
3.2 Interpretability
Based on the ‘black-box’ methods, deep learning-based AI has been questioned about the lack of interpretability which is an obstacle towards the clinical adoption of AI [81-83]. Several studies used post hoc methods or supervised ML models to explain the output of deep learning-based algorithms after DL algorithms made its prediction [47, 48]. However, post hoc analyses of DL methods have been criticized because additional models should not be required to explain how a DL model works [82]. Recently, some researchers integrated DL algorithms and hand-crafted ML approaches to raise the biological interpretability of the model. Wang et al. [84] used a DL approach to segment nuclei in digital HE images of early-stage non-small cell lung cancer to predict tumor recurrence before applying a hand-crafted method involving the interrogation of nuclear shape and texture. More strategies are needed to increase the interpretability of AI algorithms and gain confidence of doctors and patients.
3.3 Computing system
Histopathological image has large file sizes, being about 1,000 times of an X-ray and 100 times of a CT image. As a result, high specs hardware is required for both storage and processing. It is necessary to design a powerful AI model and build efficient and scalable storage and computing system to analyze the images. With the usage of cloud platforms, there are challenges in the massive bandwidth required to transmit gigapixel-sized WSI images into data clouds as well as managing permanent and uninterrupted communication channels between end-users and the cloud. These challenges would be addressed in the foreseeable future brought by improvements in information technology such as universal adoption of 5G.
3.4 Attitude of pathologists
In addition to the lack of interpretability of AI, some pathologists are afraid of the change in workflow. When AI is used, the pathologists won't observe the histopathological morphology with a microscope, but on an accelerated parallel processing (APP). How would the pathologists describe the diagnosis evidence from AI in the diagnosis report? How much responsibility should the pathologists have when they assign a diagnosis report with the help of AI? These issues need to be resolved before the real human-machine cooperation are implemented in clinical practice. In addition, with the development of AI, more and more algorithms/platforms are being developed. Another important issue for pathologists is how to choose an adaptable one and standardize the output from different algorithms/platforms.
3.5 Attitude of clinicians and patients
All of the diagnostic reports serve clinicians to develop suitable intervention programs for patients. As a result, the output of AI should be understood and trusted by the clinicians. In addition, considering the cost of patients, the clinician needs to decide the minimum diagnostic assays as well as prognostic and predictive assays. The AI-based diagnostic and prognostic/predictive assays should have a high accuracy and be convenient for routine clinical use. Similarly, AI-based tests should be trusted by patients. It is better for AI-based tests to be set up as Clinical Laboratory Improvement Amendments (CLIA)-based tests and enter into clinical guidelines.
3.6 Regulators and reimbursements
Besides the accreditation of doctors and patients, the clinical adoption of AI digital pathology needs approval by regulatory agencies. The key principle guiding the approval process in most countries is the requirement of an explanation of how the software works [85-87]. The lack of interpretability limits the approval for DL-based AI approaches [82]. Recently, the FDA has started granting approval to DL-based approaches for clinical use in the USA. Philips received an approval for a digital pathology whole-slide scanning solution (IntelliSite) in 2017 [88], and subsequently, the digital pathology solution PAIGE.AI [89] was granted Breakthrough Device designation by the FDA in 2019 [90]. Despite of the above achievements, AI-based devices tend to be assigned to Class II or III in the FDA three-class system for the approval of medical devices, in which Class I devices deem to have the lowest risk and Class III devices to have the highest risk. In the European Union, no AI solutions with prognostic/predictive intent have a Conformité Européenne marking, but the digital pathology solutions developed by Philips, Sectra and OptraSCAN have secured clearance to carry such a designation. Although the FDA apparently intends to regulate CLIA-based tests more stringently, it seems to be a better way for AI-based diagnostic assays to follow the model established by CLIA-based genomic tests to get approval for clinical usage.
Reimbursement of the costs of AI-based diagnostic and prognostic/predictive assays is one of the major issues that affect the application of these assays in clinic [91]. In the USA, insurance companies standardize expenses on the basis of the current procedural terminology codes maintained by the American Medical Association and reported by medical professionals [92-95]. At present, there are no dedicated procedure codes for the use of AI in digital pathology with diagnostic or prognostic intent. AI-based tools probably need to be approved by FDA before they get the new procedure codes and are reimbursable. In China, AI-based tools have not been covered by social medical insurance or insurance companies.
3.7 Summary
Future pathological diagnosis probably needs to incorporate multimodal measurements, such as proteomics, genomics, and measurements from multiplexed marker-staining platforms, in order to supply a comprehensive patient-specific portrait for tumor precision treatment [96]. Despite the above challenges and obstacles, the potential of DL-based AI approaches for digital pathology is promising since AI has strong feature representation learning capability enabled by improvements in algorithm, accumulation of big data and increased computing power. People will have more confidence in AI algorithms after they are validated using multi-center data and have increased interpretability. The collaboration between pathologists and AI will promote tumor precision treatment.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
COMPETING INTERESTS
The authors declare that they have no competing interests.
FUNDING
This work was partially supported by the National Nature Science Foundation of China (81871990 and 81472263).
AUTHORS’ CONTRIBUTIONS
YS and XL conceived this study. YJ, MY, XL and YS drafted the manuscript. SW revised the manuscript. All authors read and approved the final manuscript.
ACKNOWLEDGEMENTS
Not applicable.




