Spectrum of EGFR aberrations and potential clinical implications: insights from integrative pan-cancer analysis
Abstract
Background
Human epidermal growth factor receptor (EGFR) is an oncogenic gene and one of top targets of precision therapy in lung cancer with EGFR mutations. Although there are many reports for some individual cancers, comprehensive profiling of EGFR mutations, overexpression, amplification, DNA methylation, and their clinical associations across many different cancers simultaneously was not available. This study aimed to fill the gap and provide insights to the alteration spectrum of EGFR and its therapeutic and prognostic implications.
Methods
The Cancer Genome Atlas (TCGA) datasets for 32 cancer types involving 11,314 patients were analyzed for alterations (mutations and amplification/deletion), abnormal expression and DNA methylation in EGFR gene. Mutation frequency, genomic location distribution, functional impact, and clinical targeted therapy implication were compared among different cancer types, and their associations with patient survival were analyzed.
Results
EGFR alteration frequency, mutation sites across functional domains, amplification, overexpression, and DNA methylation patterns differed greatly among different cancer types. The overall mutation frequency in all cancers combined was relatively low. Targetable mutations, mainly in lung cancer, were primarily found in the Pkinase_Tyr domain. Glioblastoma multiforme had the highest rate of alterations, but it was dominated by gene amplification and most mutations were in the Furin-like domain where targeted therapy was less effective. Low-grade glioma often had gene amplification and increased EGFR expression which was associated with poor outcome. Colon and pancreatic adenocarcinoma had very few EGFR mutations; however, high EGFR expression was significantly associated with short patient survival. Squamous cell carcinoma regardless of their sites (the head and neck, lung, or esophagus) exhibited similar characteristics with an alteration frequency of about 5.0%, was dominated by gene amplification, and had increased EGFR expression generally associated with short patient survival. DNA methylation was highly associated with EGFR expression and patient outcomes in some cancers.
Conclusions
EGFR aberration type, frequency, distribution in functional domains, and expression vary from cancer to cancer. While mutations in the Pkinase_Tyr domain are more important for treatment selection, increased expression from amplification or deregulation affects more tumor types and leads to worse outcome, which calls for new treatment strategies for these EGFR-driven tumors.
Abbreviations
-
- aa
-
- amino acid
-
- ACC
-
- adrenocortical carcinoma
-
- ACMG
-
- American College of Medical Genetics and Genomics
-
- AML
-
- acute myeloid lymphoma
-
- BLCA
-
- bladder urothelial carcinoma
-
- BRCA
-
- breast invasive carcinoma
-
- CCLE
-
- Cancer Cell Line Encyclopedia
-
- CESC
-
- cervical squamous cell carcinoma and endocervical adenocarcinoma
-
- CHOL
-
- cholangiocarcinoma
-
- CNV
-
- copy number variant
-
- COAD
-
- colon adenocarcinoma
-
- DFS
-
- disease-free survival
-
- DLBC
-
- lymphoid neoplasm diffuse large B-cell lymphoma
-
- EGFR
-
- epidermal growth factor receptor
-
- ESCA
-
- esophageal carcinoma
-
- FDA
-
- Food and Drug Administration
-
- GBM
-
- glioblastoma multiforme
-
- GDC
-
- Genomic Data Commons
-
- GTEx
-
- Genotype-Tissue Expression
-
- HNSC
-
- head and neck squamous cell carcinoma
-
- ICGC
-
- the International Cancer Genome Consortium
-
- Indel
-
- insertion/deletion
-
- KICH
-
- kidney chromophobe
-
- KIRC
-
- kidney renal clear cell carcinoma
-
- KIRP
-
- kidney renal papillary cell carcinoma
-
- LGG
-
- brain lower grade glioma
-
- LIHC
-
- liver hepatocellular carcinoma
-
- LUAD
-
- lung adenocarcinoma
-
- LUSC
-
- lung squamous cell carcinoma
-
- MAPK
-
- mitogen-activated protein kinase
-
- MESO
-
- mesothelioma
-
- NSCLC
-
- non-small- cell lung cancer
-
- OS
-
- overall survival
-
- OV
-
- ovarian serous cystadenocarcinoma
-
- PAAD
-
- pancreatic adenocarcinoma
-
- PCPG
-
- pheochromocytoma and paraganglioma
-
- PI3K
-
- phosphoinositide 3-kinase
-
- PKB
-
- protein kinase B
-
- PRAD
-
- prostate adenocarcinoma
-
- RAF
-
- rapidly accelerated fibrosarcoma
-
- RAS
-
- rat sarcoma
-
- RTK
-
- receptor tyrosine kinases
-
- SARC
-
- sarcoma
-
- SKCM
-
- skin cutaneous melanoma
-
- SNV
-
- single nucleotide variant
-
- STAD
-
- stomach adenocarcinoma
-
- TCGA
-
- The Cancer Genome Atlas
-
- TGCT
-
- testicular germ cell tumors
-
- THCA
-
- thyroid carcinoma
-
- THYM
-
- thymoma
-
- TKIs
-
- tyrosine-kinase inhibitors
-
- TPM
-
- transcripts per million
-
- UCEC
-
- uterine corpus endometrial carcinoma
-
- UCS
-
- uterine carcinosarcoma
-
- UVM
-
- uveal melanoma
1 BACKGROUND
The human epidermal growth factor receptor (EGFR) family, also known as the HER family of receptor tyrosine kinases (RTK), consists of four members—EGFR, ERBB2, ERBB3, and ERBB4 [1, 2]. Five functional domains are characterized for EGFR according to the database of protein families (Pfam, http://pfam.xfam.org/protein/P00533): Recep_L (57-168aa), Furin-like (177-338aa), Recep_L (361-481aa), GF_recep_IV (505-637aa), and Pkinase_Tyr domains (712-968aa). The Recep_L domains contain ligand binding sites; the Furin-like domain is a cysteine rich region involved in signal transduction and receptor aggregation; the GF_recep_IV domain regulates the binding of a ligand to the Recep_L domains; and the Pkinase_Tyr domain performs the phosphorylation function [3].
Upon stimulation by its ligands, dimerization (both homodimerization and heterodimerization) of EGFR results in its intracellular tyrosine kinase activation and autophosphorylation at multiple tyrosine residues, which activates a number of downstream signaling cascades that not only promote proliferation, growth, and survival of normal cells but also contribute to processes that are crucial to cancer progression, including angiogenesis, metastasis, and apoptosis [4, 5]. The best known involved pathways include the rat sarcoma (Ras)/mitogen-activated protein kinase (MAPK) and phosphoinositide 3-kinase (PI3K)/protein kinase B (PKB) signaling pathways, whose roles in promoting tumor growth, survival, and progression are well characterized [6].
EGFR is one of the first few identified oncogenes and is a key treatment target in clinical oncology [7-10]. It is frequently activated by gene mutation, amplification, or overexpression through abnormal regulation in human cancers. Among EGFR-associated cancers, pancreatic adenocarcinoma (PAAD) has an extremely poor prognosis, which usually results in death within several months after diagnosis [11, 12]. In cancers like non-small-cell lung cancer (NSCLC) [13] and colon adenocarcinoma (COAD) [14], EGFR mutation status is considered as a poor prognostic factor, which is often associated with a more aggressive behavior and decreased patient survival.
Because of the critical roles of EGFR in cancers, various treatment strategies, including tyrosine kinase inhibitors (TKIs, small-molecule inhibitors, which bind to the ligand-binding site on the extracellular domain) [15], antibody-based therapy [16], immunotherapy [17], and preclinical trials of RNA interference therapies [18], have been developed to inhibit its activities and thus control tumor growth and progression. When the ligand binding with EGFR is prevented by monoclonal antibodies or TKIs, it dampens signal transduction through pathways such as the RAS/rapidly accelerated fibrosarcoma (RAF)/MAPK and PI3K/PKB cascades [2, 19]. Such treatments are very effective and provide significantly improved patient outcomes, particularly for lung adenocarcinoma (LUAD) patients with EGFR mutations [20, 21]. However, successful applications of TKIs to other cancers are less certain [22]. Although many literature reports are available on EGFR mutation, overexpression, or amplification for particular cancer types [12-14, 23, 24], a simultaneous comprehensive profiling over multiple cancer types to explore their similarity and difference is not available. Such information is important to understand what other cancers are more likely to benefit from such targeted therapy and what role EGFR plays among different cancers.
Taking advantage of the large datasets from The Cancer Genome Atlas (TCGA), we systematically profiled the mutation, copy number, expression, and DNA methylation patterns of EGFR across 32 cancer types. We first examined the patterns of EGFR mutations, including single nucleotide variant (SNV) and short insertion/deletion (indel), across tumors and their implications for targeted therapies. Copy number variants (CNVs) and their impact on gene expression and clinical outcomes were investigated next. For cancers without gene amplification, we further looked into gene expression patterns, their association with clinical outcomes, contribution of DNA methylation to gene abnormal expression, and impact of DNA methylation on patient overall survival (OS).
2 DATA SOURCES AND METHODS
2.1 EGFR mutation and CNV data from TCGA provisional dataset
Mutation data from whole exome sequencing, CNVs from GISTIC for EGFR gene, and clinical data were downloaded and formatted from the cBioportal (https://www.cbioportal.org/), a web resource for exploring, visualizing, and analyzing multidimensional cancer genomics databases, such as TCGA [25], the International Cancer Genome Consortium (ICGC) [26], Cancer Cell Line Encyclopedia (CCLE) [27]. We selected the more inclusive TCGA provisional cohort, which was retrieved on Feb 26th, 2019 consisting of 11,314 patients with 11,410 samples across 32 histopathologic cancer types and representing most major classes of human adult cancers [28, 29]. The mutation data included SNVs, indels, and CNVs (defined by GISTIC 2.0 as following for log ratio value: -2/-1 = deletion; 0 = diploid; 1 = gain; 2 = amplification).
2.2 EGFR expression and methylation data in normal and cancer tissues
EGFR expression in normal tissues was examined in RNA sequencing (RNA-seq) data from The Genotype-Tissue Expression (GTEx) (https://www.gtexportal.org/home/), which consisted of 11,688 samples from 53 tissue types/organs. Transcripts per million (TPM) was used to compare the relative expression levels of EGFR from different organs or sub-locations of an organ. Expression and DNA methylation data in tumors and their paired normal samples were obtained from TCGA Genomic Data Commons (GDC) portal (https://portal.gdc.cancer.gov/), which included data for 32 cancer types. The expression data was pre-processed and normalized using upper quartile of 1000. The genome-wide profile of human DNA methylation was generated using Infinium Human Methylation 450K BeadChip (Illumina, San Diego, CA, USA).
2.3 Data analysis
For differential expression of EGFR between tumors and their paired normal samples in each cancer type, the expression data was log2 transformed, and two-group t test was performed for those tumor types with at least two normal samples (22 out of 32 tumor types). The log2 fold change and significant P value (minus log10) were plotted by using the Volcano plot for each cancer type. As TCGA had a limited number of paired normal samples or not at all for several tumor types, we further examined the differential expression data from Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/index.html) [30], a web server for exploring and analyzing RNA-seq data of 9736 tumor samples and 8,587 normal samples from TCGA and the GTEx projects using a uniform processing pipeline and normalization method.
OS and disease-free survival (DFS), defined as the duration from diagnosis to death or recurrence, were analyzed using the Kaplan-Meier method. The hazard ratio and 95% confidence intervals were plotted by R package ‘forestplot’. Each CpG's methylation correlation with EGFR expression was analyzed using Pearson correlation. The prognostic values of EGFR alterations and its CpG methylation were analyzed with Cox proportional hazard model.
All data analyses were conducted using the R package, version 3.5.3 (https://www.r-project.org/) unless specifically stated.
3 RESULTS
3.1 EGFR somatic mutation patterns across tumor types
The overall EGFR mutation frequency was 2.8% (320/11,410) for all tumor samples and 2.4% (268/11,314) for all patients across the 32 tumor types. The most common tumors with EGFR mutations were glioblastoma multiforme (GBM, 26.8%), LUAD (14.4%), diffuse large B-cell lymphoma (DLBC, 8.3%), and skin cutaneous melanoma (SKCM, 6.5%). On the contrary, kidney chromophobe cell carcinoma (KICH), mesothelioma (MESO), pheochromocytoma and paraganglioma (PCPG), thymoma (THYM), thyroid carcinoma (THCA), uterine carcinosarcoma (UCS), and uveal melanoma (UVM) showed almost no EGFR mutations (Figure 1A). The total number of samples for each cancer type varied from 48 (DLBC) to 1105 (breast invasive carcinoma [BRCA]), and those with too few samples might not reflect the complete picture of EGFR mutation status (Supplementary Table S1).
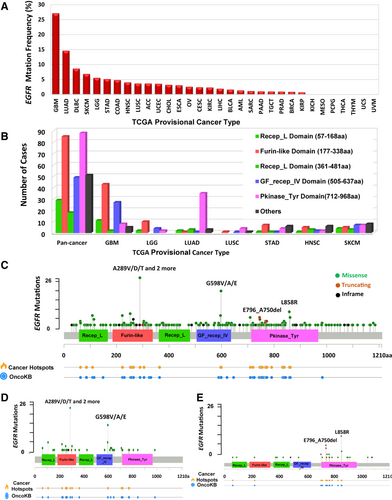
The 320 EGFR somatic mutations (from 268 tumor samples) were observed across all cancer types and were widely distributed along different functional domains of EGFR gene. The most common ones were the Pkinase_Tyr domain (88 samples) and the Furin-like domain (85 samples), followed by the GF_recep_IV domain (45 samples). The location distribution of these EGFR mutations was dramatically different among different cancers (Figure 1B, Supplementary Table S2). Mutations in GBM and brain lower-grade glioma (LGG) were most commonly located in the Furin-like domain, about 5 times more than the mutations located in the Pkinase_Tyr domain. On the contrary, mutations in NSCLC were primarily in the Pkinase_Tyr domain, especially for LUAD, which amounted to four fifths of all mutations. Mutations in stomach adenocarcinoma (STAD), head and neck squamous cell carcinoma (HNSC), and SKCM were mostly in other domains whose functions were less known.
From functional impact on protein coding, these 320 EGFR mutations were classified into three categories: missense (280 mutations), truncating (21 mutations), and in-frame insertion/deletions (19 mutations) (Figure 1C). The 289aa in the Furin-like domain was the most frequently mutated position, which was observed in 27 samples (3 samples with A289D, 1 with A289I, 1 with A289N, 6 with A289T, 15 with A289V, and 1 with A289Rfs*9). The mutations at this position almost exclusively occurred in GBM samples (25/27) (Figure 1D). A289V is known to be oncogenic, while other mutation types (A289D/T/N/I) are likely oncogenic. None of these mutations are known to be targetable. The only other tumor with mutations at this position was HNSC (1 sample with A289T and 1 with A289Rfs*9), and their importance was little known to this cancer. The second most mutated position was 598aa in the GF_recep_IV domain: 16 GBMs had G598V, 2 GBMs had G598A, and 1 esophageal squamous cell carcinoma (ESCC) had G598E. These mutations might affect ligand-receptor binding and are likely oncogenic. Most mutations in LUAD (35 of 45 mutations) were located in the Pkinase_Tyr domain, especially at the positions of 858aa (8 samples with L858R) and 746-750aa (6 with E746_A750del, 2 with L747_E749del, and 1 with L747_T751del) (Figure 1E).
Each somatic gene mutation can be classified by their oncogenic effect and predictive significance [31]. As shown in Figure 2, 83 (25.9%) EGFR mutations were oncogenic, 34 (10.6%) likely oncogenic, 27 (8.4%) predicted oncogenic, 5 (1.6%) likely neutral, 3 (0.9%) inconclusive, and 168 (52.5%) unknown. Over half of them were in the unknown class, highlighting the challenge in mutation interpretation.
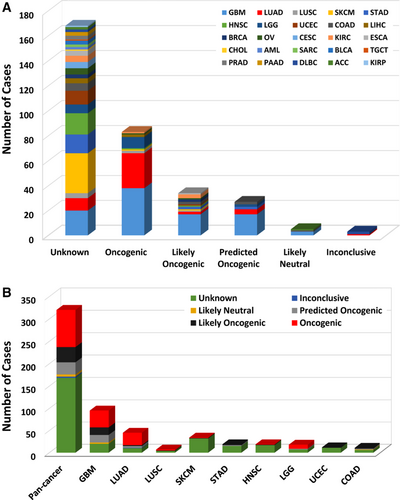
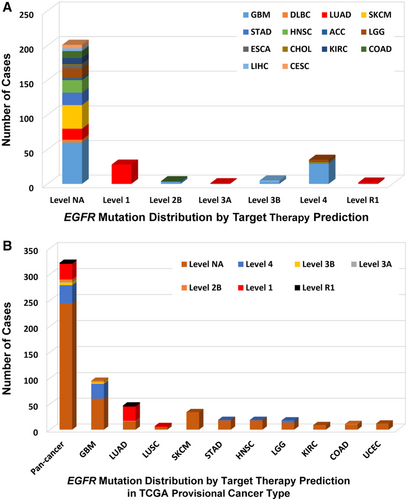
According to clinical targeted therapy implication [32], the 320 EGFR mutations could be divided into 7 levels (Supplementary Table S1): Level 1 (30 mutations), Level 2B (5 mutations), Level 3A (1 mutation), Level 3B (5 mutations), Level 4 (33 mutations), Level NA (244 mutations), and Level R1 (2 mutations). Only Level 1 and Level R1 mutations were indicated for or against targeted therapy currently with a Food and Drug Administration (FDA)-approved drug [33]. All Level 1 mutations were found in NSCLC (28 in LUAD and 2 in lung squamous cell carcinoma [LUSC]), and these mutations were concentrated in exons 19-21, which included L858R, L861Q, G719A, S768I, L833F, E796_A750del, L747_E749del, E709_T710delinsD, L747_T751del, and T751_E758del (Figure 3). LUAD harbored the highest proportion of oncogenic/likely oncogenic mutations (66.7%), and almost all were Level 1 mutations (28 of 30 mutations). Although the oncogenic/likely oncogenic mutations accounted for 57.9% and 50.0% of GBM and LGG samples, respectively, most of them were in Level 4 and Level NA without treatment implications.
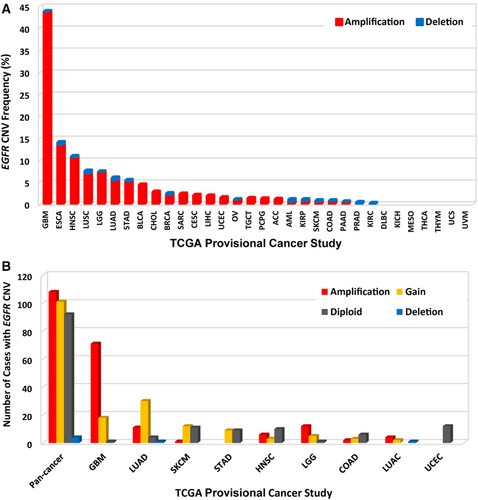
3.2 EGFR CNVs in different tumor types
The overall EGFR CNV frequency was about 5.0% (detected in 569 of 11,410 samples). Almost all were amplification (in 549 of 569 samples), only 20 samples with deletion (Figure 4A). The most common solid tumors with EGFR CNVs were GBM (43.9%), ESCA (14.1%), HNSC (10.9%), LUSC (7.6%), and LGG (7.4%). DLBC, kidney renal clear cell carcinoma (KIRC), MESO, THCA, THYM, UCS, and UVM were not affected with any CNV (Figure 4A). Among the 320 samples with EGFR mutations described above, 213 also had EGFR CNV changes (Figure 4B, Supplementary Table S1), of which 108 with amplification, 101 with gain, and 4 with deletion. GBM and LUAD were two cancer types with the highest numbers of amplification or gain (Figure 4B).
3.3 Combined EGFR alterations (mutation and CNV) in different cancer types
The combined EGFR mutation and CNV frequency in all tumors was about 7.0% (746 of 11,314 patients, 748 of 11,410 samples). However, the frequency among different cancers was dramatically different (Figure 5A). While KICH, MESO, THCA, THYM, USS, and UVM had neither EGFR mutation nor EGFR CNV, 273 of 591 (46.2%) GBM cases had CNV (33.2%), mutation (3.4%), or both (9.6%). Other cancers with dominantEGFR amplification but at much lower amplification rate included ESCA (13.0%), HNSC (9.4%), STAD (5.2%), LGG (5.4%), LUSC (6.4%), and BLCA (4.4%). Mutation was more common in DLBC (8.3% vs. 0.0%) and SKCM (5.2% vs. 0.2%). Although EGFR alteration frequencies of two lung cancer subtypes (LUAD and LUSC) were similar, the mutation frequency was slightly higher than that of amplification in LUAD (4.7% vs. 3.7%), while amplification was much more common than mutation in LUSC (6.4% vs. 0.6%).

Mutation location and CNV occurrence appeared to be associated. Over half of the mutations (47 of 82 mutations) in the Furin-like domain were accompanied by EGFR amplification, while nearly half of mutations (40 of 85 mutations) in the Pkinase_Tyr domain had copy gain. Mutations in the Recep_L domains and other function-unknown domains rarely had concurrent CNVs (Figure 5B).
3.4 EGFR alterations and patient survival
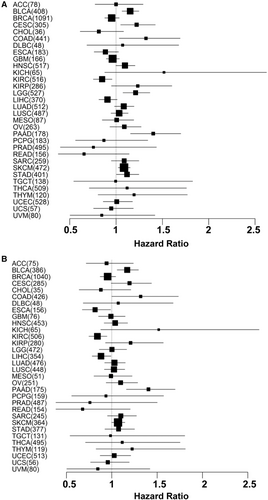
In order to evaluate the clinical significance of EGFR alterations, we analyzed patient survival for pan-cancer and for each cancer type separately by alteration status (mutations and CNVs alone or in combination). When all tumors were analyzed together, patients with any EGFR alteration had significantly shorter median OS and DFS than those without EGFR alteration (both P < 0.001, Supplementary Figure S1). When analysis was performed for CNV and mutation separately, the presence of either aberration was associated with shortened patients’ OS and DFS (all P < 0.001, Supplementary Figure S2 and S3).
For survival association in individual cancer types, only those cancer types with at least 10 tumor samples containing either EGFR mutations or CNVs were included in the analysis. Among patients with HNSC, LGG, or LUAD, EGFR amplification was associated with short survival (Figure 6A). However, EGFR somatic SNV/indel mutations appeared not affecting patient survival although only 5 cancer types with at least 10 samples containing EGFR mutations could be analyzed (Figure 6B). This low mutation frequency might lead to an insufficient power to detect the small effect of mutations on survival. Not surprisingly, EGFR-amplified tumors had significantly higher EGFR expression than those without EGFR amplification in all 9 cancers types (all P < 0.001, Figure 6C); however, there was no much EGFR expression difference between tumors with or without EGFR mutations except that EGFR mutation status was associated with significantly increased EGFR expression in GBM (P = 0.024) and LUAD (P = 0.001, Figure 6D).
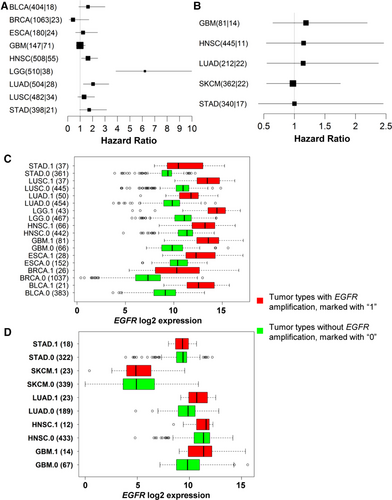
3.5 EGFR abnormal expression and clinical implications
The low EGFR mutation frequency in most cancer types made it difficult to assess its impact on patient survival yet, and the increased expression of EGFR wide type could be oncogenic driver for tumor development and progression and have a negative impact on patient survival. For this we were interested in the tumors with abnormal EGFR expression compared with their paired normal samples across different cancer types. We first examined EGFR expression in 53 types of normal tissues in GTEx samples. As shown in Figure 7A, the expression of EGFR was quite variable across different types of tissues, with the medians ranging from 2.22 TPM (the brain-spinal cord) to 73.91 TPM (the skin on the sun-exposed, lower leg). EBV-transformed lymphocytes and whole blood virtually barely had EGFR expression. The skin (both sun-exposed and not exposed) had the highest expression.
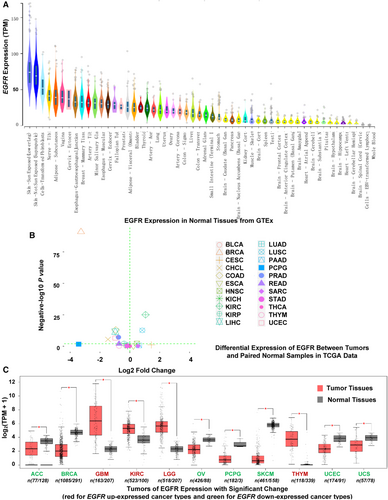
We then compared EGFR expression between tumors and their paired normal samples profiled in TCGA. Among the 32 tumor types, 22 had at least two normal samples. Differential expression was found in 10 cancer types (all P < 0.01), with 3 cancer types up-regulated (HNSC, LUSC, and KIRC) and 7 down-regulated (BRCA, cholangiocarcinoma [CHOL], uterine corpus endometrial carcinoma [UCEC], liver hepatocellular carcinoma [LIHC], COAD, prostate adenocarcinoma [PRAD], and kidney renal papillary cell carcinoma [KIRP]) (Figure 7B). The most noticeable was BRCA whose EGFR expression was reduced over 8 folds (log2 fold change = -3). The cancer type with the most increased expression was KIRC with over 2-fold increase. As some cancer types in TCGA did not have paired normal tissues or had too few, we further analyzed the data using GTEx normal samples as reference. Among different cancer types, GBM, LGG, KIRC, HNSC, ESCA, and LUSC all had higher EGFR expression than other cancer types (Supplementary Figure S4). Differential expression analysis from this expanded normal reference dataset showed similar results to the analysis of TCGA dataset. However, it revealed several additional cancer types with EGFR differential expression. EGFR expression was up-regulated in GBM, LGG, KIRC, and THYM but down-regulated in adrenocortical carcinoma (ACC), BRCA, ovarian serous cystadenocarcinoma (OV), PCPG, SKCM, UCEC, and UCS (all P < 0.01, Figure 7C). In the normal brain tissues, EGFR had minimal expression; however, its expression in GBM and LGG was dramatically increased (20-fold increase for GBM and more than 10-fold increase for LGG). KIRC also had nearly 4-fold increase as compared with normal kidney tissues. Both ESCA and LUSC had a higher level of EGFR expression (about 2 folds) than their matching normal tissues. EGFR expression in SKCM was dramatically reduced relative to normal skin tissues; however, SKCM is thought to originate from melanocytes, and normal skin tissues have few such cells.
Survival association analysis using all tumors regardless of mutation or CNV status showed that increased EGFR expression was associated with short patient OS in bladder urothelial carcinoma (BLCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), COAD, HNSC, LGG, PAAD, SKCM, and STAD. Interestingly, increased EGFR expression was associated with better prognosis for KIRC (Figure 8A). As gene amplification is a major driver for increased expression, we then limited analysis to those samples without amplification (tumors with unknown CNV status were also excluded). The same association held for BLCA, CESC, COAD, PAAD, SKCM, and KIRC (Figure 8B).
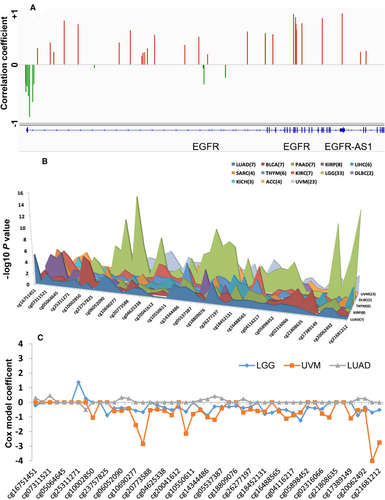
3.6 Associations of CpG methylation with EGFR expression and patient survival
For the tumors without EGFR amplification, we were interested in knowing if DNA methylation changes were responsible for or associated with abnormal expression of EGFR. Therefore, we extracted the methylation data of the 49 CpG sites associated with EGFR for the same set of patients. We analyzed the associations of each CpG site with EGFR RNA expression for all tumors together (7,913 samples) and for each tumor type separately. Among the 49 CpG sites, 46 were significantly associated with EGFR expression after Bonferroni multiple testing correction (P < 0.001). Interestingly, all CpGs around the promoter region of EGFR (7 CpGs in TSS+/-1500 or 5′ UTR region) were negatively correlated with EGFR expression, while almost all (except 4) in the gene body were positively correlated with EGFR expression (Figure 9A). In individual cancer type analysis, most tumor types had the similar patterns of methylation association with gene expression in promoter and gene body, but a few others had predominant hypomethylation in both regions, such as GBM, LUSC, PRAD, THYM, KIRC, and KICH (Supplementary Figure S5). Survival analysis for all tumors with tumor type as a covariate only found 2 CpG sites (cg07311521 and cg16751451) significantly associated with OS, and both were in the promoter region (TSS1500). Association analysis for each tumor type separately found 13 tumor types with at least 2 CpGs that were significantly associated with OS (Figure 9B). LGG and UVM had the highest numbers of CpGs associated with patient survival (33 and 23 CpGs, respectively). For both cancer types, the CpGs with significant associations were mostly located in the gene body, where higher CpG methylation was associated with a better outcome. Higher methylation of one CpG in the promoter region was significantly associated with worse outcome of LGG. However, for LUAD (with 7 significant CpGs), their associations were mostly in opposite directions: hypermethylation of CpGs in the gene body was associated with worse outcome, while hypermethylation of one CpG in the promoter region was associated with better outcome (Figure 9C).
4 DISCUSSION
Our analyses showed that EGFR mutation frequency, locations, and amplification differed greatly among different cancer types, which had important clinical implications. GBM had the highest rate of EGFR alterations, and amplification was the primary alteration. Somatic mutations generally occurred in the non-targetable Furin-like domain. Paradoxically, although EGFR expression in GBM was significantly increased, either by gene amplification or DNA methylation change, the expression level was not associated with patient OS. Other common tumor types with EGFR alterations include ESCA, HNSC, LGG, LUSC, and BLCA, all with similar characteristics: alteration frequency of about 5.0% and amplification as a dominant type. Except LGG, they were squamous or similar transitional cell origin. LUAD and STAD had similar frequencies of alternations, but their mutation and amplification patterns differed. On the other side of the spectrum, tumors such as DLBC and SKCM mainly had SNV mutations but rarely CNV; tumors including KIRC, MESO, THCA, THYM, UCS and UVM almost had no EGFR alterations.
Mutations in the Furin-like and Pkinase_Tyr domains accounted for most of EGFR single nucleotide or indel mutations. However, the Pkinase_Tyr domain was far more important in terms of targeted therapy with TKIs as 90% EGFR mutations in LUAD occurred in this region, particularly the exon 19 deletion and the L858R point mutation in exon 21. Mutations in these regions are proven predictive markers for effective TKI therapy for NSCLC in clinical practice [7, 33-35], with significantly prolonged survival as compared with traditional combination chemotherapy [21, 36, 37]. The mutation rate of NSCLC in this TCGA cohort appeared consistent with the previous report that these two mutations accounted for 85% to 90% of all EGFR-mutated NSCLC tumors [38, 39]. For other uncommon EGFR mutations in NSCLC, targeted therapy generated inconsistent results [34, 40-42]. It is clear that different EGFR mutations have very different implications, and only those resulting in destabilization of the equilibrium between the active and inactive states of EGFR kinase activity may benefit from EGFR targeted therapy [43, 44].
The clinical significance of EGFR mutations in other regions of the gene is less defined. EGFR was first linked to the oncogenesis of GBM [45]. In this large TCGA dataset, the combined alteration rate (amplification, deletion, or mutation) reached 67.3% in GBM. However, this high alteration rate was mainly driven by a high frequency of gene amplification. Compared with LUAD, most EGFR mutations in GBM were located in extracellular domain or single-span transmembrane segment, which was known to be associated with tumorigenesis but not responsiveness to TKIs. Although EGFR amplification was a predictor of poor prognosis for several cancer types, it was not significantly associated with GBM, consistent with the paradox phenomenon reported in the literature [46]. Similarly, EGFR expression level was not prognostic in this dataset although some reports suggested otherwise [47]. The disagreement could be the results of multiple factors. The absence of well-known responsive mutations and presence of redundant and alternative compensatory pathways were among the most important escape mechanisms [48].
The prognostic role of EGFR in LGG appears clear. Both amplification and high expression of EGFR were correlated with short patient survival in this dataset as reported previously [49, 50]. More interestingly, we found that LGG had the highest number of CpGs whose methylation level was associated with patient survival (i.e., hypermethylation of CpGs in the gene body with better survival), which has not been reported before.
COAD and PAAD had very few EGFR mutations in this TCGA dataset. However, high EGFR expression was significantly associated with short patient survival. Studies have shown that inhibition of EGFR by TKIs or antibodies either alone or in combination with chemotherapy provided extended survival for patients with these cancers [51-53]. Unfortunately, the clinical benefit is generally not big enough for routine application.
Squamous cell carcinomas in the head and neck (HNSC), lung (LUSC), and esophagus (ESCA) have some commonalities: significantly increased EGFR expression, high frequency of EGFR amplification, and low rate of SNV/indel mutations. Targeted therapy with cetuximab or necitumumab (a monoclonal antibody targeting EGFR), along with radiotherapy or chemotherapy, have demonstrated promising efficacy and prolonged OS for locally advanced or recurrent and/or metastatic HNSC [54], ESCA [55-57], and LUSC [58, 59], which is now a new first-line treatment option in squamous NSCLC. The clear correlation between EGFR abnormal expression and treatment benefit highlights the importance of molecular profiling and predictive biomarkers for treatment selection.
This study profiled 32 cancer types. However, some rare cancer types did not have sufficient samples to capture the full EGFR alteration and expression spectrum and establish small and moderate associations. The low frequency of EGFR mutation or amplification also made analysis challenging. It was mostly a pan-cancer global survey without deep dive on each cancer type. Several important leads revealed from this study are the directions of our future investigations.
5 CONCLUSIONS
Our analysis provides a comprehensive view of EGFR mutation, abnormal expression, DNA methylation, and their interplay and clinical implications for 32 cancer types covering over ten thousand tumor samples. While some alternations are involved more in tumorigenesis, others are more therapeutic. Some cancer types have a higher frequency of EGFR alternations where mutation, amplification, or abnormal expression is associated with outcome or indicated for clinical action. Genomic profiling may provide guidance for their use in targeted therapy.
DECLARATIONS
ACKNOWLEDGEMENTS
The results shown here are in whole or part based upon data generated by The Cancer Genome Atlas (TCGA) Research Network: https://www.cancer.gov/tcga.
FUNDING
This work was supported by China Scholarship Council (No. 201806015028 to HJL), Chinese National Natural Science Foundation (No. 81101998 to HJL, No. 81872018 and No. 81372292 to BZ), Key Project from the Chinese Ministry of Science and Technology (No. 2017YFC0110200 to BZ), and the Mayo Clinic Center for Individualized Medicine (to ZFS).
AUTHORS’ CONTRIBUTIONS
HJL and ZFS conceived the study, performed the data analyses, interpreted the results, and wrote the manuscript. BZ provided suggestions and feedbacks to the design and implementation of the study. All authors read and approved the final manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AVAILABILITY OF DATA AND MATERIALS
The datasets analyzed for this study are all publicly available as indicated in the Method section.




