Association of HLA-G 3′-UTR Haplotypes With Recurrent Spontaneous Abortion in Women From Northwest Iran
Funding: The authors received no specific funding for this work.
ABSTRACT
Background and Objective(s)
Human leukocyte antigen-G (HLA-G) is a critical protein in immune regulation and tolerance. Recurrent spontaneous abortion (RSA) is a complex disease influenced by genetic, immune dysfunction, and environmental factors. This study investigates the role of HLA-G polymorphisms in the development of RSA.
Methods
Blood samples were collected from 80 women with RSA and 200 women without a history of RSA. After DNA extraction, PCR was used to sequence the 3′-UTR region. Allelic and genotypic frequencies were analyzed, and Haploview software was used for haplotype analysis.
Results
Individual polymorphisms did not significantly differ between the two groups. However, haplotype analysis revealed significant differences. The UTR-2 haplotype was more frequent in the RSA group compared to the healthy control group (p = 0.020), suggesting a potential association. Conversely, the UTR-4 haplotype had a significantly lower frequency in the RSA group (p = 0.041), indicating a protective role against RSA.
Conclusions
While individual polymorphisms did not differ significantly, haplotype analysis identified significant associations with RSA. These findings provide valuable insights into the genetic basis of the disease and may contribute to the development of new treatments and diagnostic tools.
1 Introduction
Recurrent spontaneous abortion (RSA) is defined as three consecutive pregnancy losses before 20 weeks from the last menstrual period. As stated by the European Society of Human Reproduction and Embryology, two or more abortions should be considered for the diagnosis of RSA. Currently, however, it is believed that patients who struggle with two consecutive abortions share a risk of RSA with those who experience three consecutive abortions (Bender Atik et al. 2018; Moqadami et al. 2023). Around 1%–2% of fertile women globally are affected by it (Ford and Schust 2009). As the rate of abortions rises, there is a decreasing prevalence of fetal chromosomal abnormalities, which are seen in 29%–60% of women with abortions, indicating that pregnancy loss may be caused by other processes (Ogasawara et al. 2000). Endocrine problems must be treated, and the most prevalent autoimmune cause of pregnancy difficulties, antiphospholipid syndrome (APS), can be adequately managed to prevent abortion (Larsen et al. 2013). Despite the fact that RSAs may have a definite etiology, such as uterine structural malformations, chromosome anomalies, hormonal imbalances, and blood system diseases, over 60% of the triggers of RSA remain unknown, with the majority of them thought to be connected to immunological abnormalities (Banadakoppa et al. 2014; Jaslow, Carney, and Kutteh 2010; Sha et al. 2017). The semi-allogeneic fetal tissue must be able to be accepted by the mother's immune system during pregnancy. Because of this, several mechanisms interact at the fetal–maternal interface (Than et al. 2019).
Around 2.5% of chromosome 6 comprises the human leukocyte antigen (HLA) region, a 3.6-Mb high-density gene located at 6p21.3 that contains approximately 200 genes (Klein and Sato 2000; Najafi et al. 2020). HLA-G is recognized as a crucial molecule that plays a significant role in establishing fetal-induced maternal immunological tolerance. Eliminating HLA class I facilitates T and B cell resistance against allorecognition I of antigens A and B and HLA class II on embryonic trophoblast cells; nevertheless, the existence of HLA-C, E, F, G provides self-signals to regulate natural killer (NK) reactions (Ishitani et al. 2003; Kovats et al. 1990). Low HLA-G levels have been related to RSA (Ishitani et al. 2003; Kofod et al. 2017). By alternative splicing, the HLA-G pre-mRNA may generate seven different isoforms: three of these are soluble (HLA-G5, -G6, and -G7), while four of these are membrane-bound (HLA-G1, -G2, -G3, and -G4) (Lee et al. 1995). Particularly, soluble HLA-G is present in amniotic fluid, blood, and seminal plasma in addition to membrane-bound HLA-G, which is predominantly expressed on trophoblasts in healthy tissue (Larsen et al. 2011; Rebmann et al. 1999; Rizzo et al. 2013; Shaikly et al. 2008). A procedure that yields soluble HLA-G1 is the cleavage of membrane-bound HLA-G from the cell surface by metalloproteinases (Rizzo et al. 2013).
The 3′ untranslated region (3′-UTR) of the HLA-G gene is highly variable. Even though the 3′-UTR is a noticeably short region, at least nine polymorphisms of this location are studied worldwide (Michita et al. 2016). The stability of mRNA is modified by the presence of 14-bp insertion /deletion regions in the 3′-UTR of HLA-G (rs371194629) (Kalotra et al. 2018). Additionally, the +3142C/G (rs1063320) sites enhance a particular microRNA's (miRNA) affinity for the HLA-G mRNA and reduce the expression of the HLA-G gene (Guberina et al. 2017; Porto et al. 2015). On the other hand, proximal AU-rich alteration at the +3187A/G (rs9380142) locations affects mRNA stability (Bai et al. 2022). There are, however, some practical investigations on single-nucleotide polymorphisms (SNPs) that are considered to be prospective miRNA binding sites, such as the +3003T/C (rs1707), +3010G/C (rs1710), +3027C/A (rs17179101), +3035C/T (rs17179108), and +3227G/A (rs1233331).
Although previous studies have demonstrated the significance of HLA-G 3′-UTR expression in individuals with RSA (Amodio et al. 2016; Hashemi et al. 2017; Meuleman et al. 2018), the relevance of the Azerbaijani population still needs to be discovered. To the best of our knowledge, this research is the first effort to investigate the importance of primary and secondary RSA in HLA-G 3′-UTR polymorphism and haplotype structure in the Azerbaijani population.
2 Materials and Methods
This case–control study was conducted from 2020 to 2023 on women referring to the “Madar” Infertility Center in Tabriz City (East Azerbaijan province, Iran). Informed consent was obtained from the volunteers participating in this study, and their information was recorded. This study protocol followed the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Medical Ethics Committee of the University of Tabriz (IR.TABRIZU.REC.1398.029). In this study, 80 women with a history of RSAs (at least two abortions), who had no known anatomical or hormonal reasons for their abortions and did not have any other diseases, were considered as the patient group, and 200 other women without a history of abortions were considered as the control group. The people of the two groups were matched in terms of age and lack of underlying disease; 4 mL peripheral blood samples were obtained from 80 patients with an age range of 20–45 years and 200 control subjects with an age range of 23–43 years. Blood samples were stored in sterile tubes containing EDTA in a freezer at −20°C for DNA extraction.
2.1 DNA Extraction and Polymerase Chain Reaction
The salting out method was employed to extract DNA. The quality and quantity of the extracted DNA were evaluated using a 1% agarose gel and spectrophotometry. Subsequently, the obtained DNA samples were amplified using the PCR technique. PCR reactions in the final volume of 40 μL include 1 μL of DNA (500 ng), 0.4 μL of Taq polymerase (5 units/μL), 4 μL of PCR buffer (10×), 0.8 μL of dNTPs (10 mM), 0.8 μL of each of the forward and reverse primers (10 nM), 1.4 μL of MgCl2 (50 mM), and 30.8 μL of double distilled water were used. The sequence of forward and reverse primers was F: ATGTATTGAGCATGTGATGGG and R: GATAACACAGGAACTTCTACGTG. The primers were designed using Oligo7 software, and their specificity was checked by Primer-BLAST online software and Gene Runner software (version 6.5.5). The PCR process was carried out in a thermocycler, including initial denaturation for 1 cycle of 5 min at 95°C, followed by 40 cycles of 30 s of annealing at 58°C, 30 s of extension at 72°C and followed by 5 min of final extension at 72°C. The amplified products were detected by electrophoresis on Green viewer-stained 2% agarose gels.
2.2 Sequencing and Genotyping
After confirming the specificity and observing a specific band in all samples, the PCR products were sent to Microsynth in Switzerland for sequencing. The sequencing Chromas software (version 2.6.6) was used to check the results.
2.3 Statistical Analysis
Allelic and genotypic frequencies in the studied population were calculated using SPSS statistical software (version 26) and GraphPad Prism software (version 8.3.0). To compare frequencies between two groups, Fisher's exact test was used to estimate the association of alleles and genotypes with the disease from the odds ratio (OR), and Haploview software (version 4.2) was used to check the haplotypes of SNPs. A p-value less than 0.05 was considered significant.
3 Results
Patients had a mean age of 32.6 ± 4.2 years, while the control group, consisting of age-matched women, had a mean age of 32.6 ± 5.1 years. Among the patients, there were 162 spontaneous abortions out of 206 pregnancies (78.6%), and 41 of the 206 pregnancies resulted in live births (20%). Additionally, 44 out of 80 patients (55%) reported a family history of abortion (see Table 1).
| Patients | Controls | |
|---|---|---|
| Total | 80 | 200 |
| Age (mean ± SD) | 32.6 ± 4.2 | 32.6 ± 5.1 |
| Spontaneous abortion (n, %) | 162/206 (78.6%) | 0 |
| Intrauterine fetal death (n, %) | 3/206 (1.4%) | 0 |
| Live birth (n, %) | 41/206 (20%) | 200 (100) |
| Family history of abortion | ||
| Yes (%) | 44 (55) | 0 (0) |
| No (%) | 36 (45) | 200 (100) |
After DNA extraction, a 675-bp fragment was amplified using PCR. The specificity of the amplification was confirmed through 2% agarose gel electrophoresis.
Table 2 shows the distribution of different genotypes and alleles of rs371194629, rs1707, rs1710, rs17179101, rs17179108, rs1063320, rs9380142, and rs1610696 in the 3′-UTR region of the HLA-G gene in patient and control groups. There were no significant differences between the allele frequencies and genotypes in the case of polymorphisms. As shown in Figure 1, there is a linkage disequilibrium between eight pairwise polymorphisms in the 3′-UTR region of the HLA-G gene. The significant D′ value is denoted by red, while a low D′ value is indicated by white.
| HLA-G | Patients (n = 80) | Controls (n = 200) | p-value | OR (95% CI) |
|---|---|---|---|---|
| rs371194629 (+2960) | ||||
| D | 109 (68.12) | 256 (64%) | 0.40 | 1.20 (0.81–1.78) |
| I | 51 (31.88) | 144 (36%) | ||
| DD | 45 (56.25) | 93 (46.5%) | 0.18 | 1.48 (0.88–2.49) |
| DI | 19 (23.75) | 70 (35%) | 0.09 | 0.58 (0.32–1.05) |
| II | 16 (20) | 37 (18.5%) | 0.67 | 1.22 (0.63–2.36) |
| rs1707 (+3003) | ||||
| T | 132 (82.5) | 340 (85%) | 0.54 | 1.20 (0.74–1.97) |
| C | 28 (17.5) | 60 (15%) | ||
| TT | 56 (70) | 144 (72%) | 0.85 | 0.91 (0.51–1.60) |
| TC | 20 (25) | 52 (26%) | 0.98 | 0.95 (0.52–1.72) |
| CC | 4 (5) | 4 (2%) | 0.33 | 2.58 (0.63–10.57) |
| rs1710 (+3010) | ||||
| G | 71 (44.38) | 174 (43.5%) | 0.92 | 1.04 (0.72–1.50) |
| C | 89 (55.62) | 226 (56.6%) | ||
| GG | 22 (27.5) | 42 (22%) | 0.31 | 1.43 (0.79–2.59) |
| GC | 27 (33.75) | 90 (45%) | 0.11 | 0.62 (0.36–1.07) |
| CC | 31 (38.25) | 68 (34%) | 0.98 | 1.05 (0.61–1.80) |
| rs17179101 (+3027) | ||||
| C | 140 (87.5) | 356 (89%) | ||
| A | 20 (12.5) | 44 (11%) | 0.72 | 1.16 (0.66–2.03) |
| CC | 63 (78.75) | 157 (78.5%) | 0.90 | 1.02 (0.54–1.91) |
| CA | 14 (17.5) | 42 (21%) | 0.62 | 0.80 (0.41–1.56) |
| AA | 3 (3.75) | 1 (0.5%) | 0.13 | 7.56 (0.77–73.74) |
| rs17179108 (+3035) | ||||
| C | 130 (81.25) | 316 (79%) | 0.33 | 1.22 (0.82–1.83) |
| T | 30 (18.75) | 84 (21%) | ||
| CC | 57 (71.25) | 142 (71%) | 0.59 | 0.87 (0.52–1.46) |
| CT | 16 (20) | 32 (16%) | 0.82 | 0.93 (0.48–1.80) |
| TT | 7 (8.75) | 26 (13%) | 0.35 | 1.37 (0.71–2.67) |
| rs1063320 (+3142) | ||||
| G | 93 (58.12) | 232 (58%) | 0.98 | 1.41 (0.99–2.00) |
| C | 67 (41.88) | 168 (42%) | ||
| GG | 33 (41.25) | 73 (36.5%) | 0.54 | 1.22 (0.72–2.08) |
| GC | 27 (33.75) | 86 (43%) | 0.19 | 0.68 (0.39–1.16) |
| CC | 20 (25) | 41 (20.5%) | 0.50 | 1.29 (0.70–2.38) |
| rs9380142 (+3187) | ||||
| A | 127 (79.38) | 304 (76%) | 0.45 | 1.22 (0.78–1.90) |
| G | 33 (20.62) | 96 (24%) | ||
| AA | 53 (66.25) | 120 (60%) | 0.40 | 1.31 (0.76–2.25) |
| AG | 21 (26.25) | 64 (29%) | 0.42 | 0.76 (0.42–1.35) |
| GG | 6 (7.5) | 16 (8%) | 0.91 | 0.93 (0.35–2.48) |
| rs1610696 (+3196) | ||||
| C | 115 (71.88) | 276 (69%) | 0.57 | 1.15 (0.77–1.72) |
| G | 45 (28.12) | 124 (31%) | ||
| CC | 43 (53.75) | 104 (52%) | 0.89 | 1.08 (0.64–1.80) |
| CG | 29 (36.25) | 68 (34%) | 0.83 | 1.10 (0.64–1.90) |
| GG | 8 (10) | 28 (14%) | 0.48 | 0.68 (0.30–1.57) |
- Note: D indicates a 14 bp deletion allele, and I indicate a 14 bp insertion allele. D = deletion; I = insertion. Further letters are nucleotides.
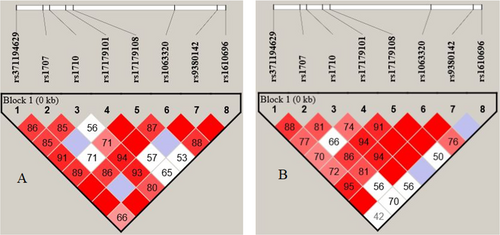
After defining the genotypes of patients and controls using Haploview software, several haplotypes were identified, as detailed in Table 3. Among these haplotypes, UTR-2 was found to be more prevalent in the RSA group compared to the healthy control group, with statistical significance (p = 0.02). This haplotype is considered a risk factor for RSA, with an OR of 1.78 and a 95% confidence interval (CI) of 1.12–2.84. In contrast, the UTR-4 haplotype exhibited a lower frequency in the RSA group, indicating a statistically significant difference (p = 0.04), which suggests its protective role against RSA. No significant differences were observed between the case and control groups for other haplotypes.
| dbSNP | rs371194629 | rs1707 | rs1710 | rs17179101 | rs17179108 | rs1063320 | rs9380142 | rs1610696 | RSA (2n = 160) | Healthy sample (2n = 400) | p-value | OR (95% CI) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HLA-G position | 2960 | 3003 | 3010 | 3027 | 3035 | 3142 | 3187 | 3196 | ||||
| UTR-1 | Del | T | G | C | C | C | G | C | 0.20 | 0.23 | 0.43 | 0.81 (0.52–1.28) |
| UTR-2 | Ins | T | C | C | C | G | A | G | 0.22 | 0.14 | 0.02* | 1.78 (1.12–2.84) |
| UTR-3 | Del | T | C | C | C | G | A | C | 0.16 | 0.12 | 0.30 | 1.36 (0.81–2.27) |
| UTR-4 | Del | C | G | C | C | C | A | C | 0.08 | 0.15 | 0.04* | 0.50 (0.27–0.94) |
| UTR-5 | Ins | T | C | C | T | G | A | C | 0.05 | 0.04 | 0.98 | 1.12 (0.47–2.66) |
| UTR-7 | Ins | T | C | A | T | G | A | C | 0.08 | 0.07 | 0.87 | 1.12 (0.58–2.18) |
| UTR-10 | Del | T | C | C | C | G | A | G | 0.01 | 0.04 | 0.22 | 0.41 (0.12–1.41) |
| UTR-15 | Ins | T | C | C | C | G | A | C | 0.04 | 0.02 | 0.29 | 1.98 (0.71–5.55) |
| UTR-18 | Del | T | G | C | C | C | A | C | 0.03 | 0.01 | 0.25 | 2.28 (0.72–7.16) |
| Others | 0.09 | 0.13 |
- Note: Significant p values are shown in bold.
4 Discussion
HLA-G is widely known for its ability to modulate the immune system, and extensive research has been conducted regarding its expression in relation to reproductive health. Polymorphisms in the 3′-UTR can impact miRNA binding sites, leading to alterations in gene expression levels. Dysregulation of gene expression can contribute to the development or progression of various diseases, including cancer, cardiovascular disorders, neurological conditions, and autoimmune diseases (Alwan Chyad et al. 2024; Arnaiz-Villena et al. 2021; Gomes et al. 2020; Haghi et al. 2021; Poomarimuthu et al. 2017). Evidence suggests that anomalies in HLA-G expression or specific variations in the HLA-G gene might be associated with RSA (Chen et al. 2022). The underlying mechanisms behind these associations are complex and likely involve interactions between the maternal immune system and the developing embryo. Notably, significant scientific attention has been given to the 3′-UTR polymorphisms of the HLA-G gene in the context of reproductive health, specifically in cases of RSA. Researchers have investigated particular variations in the 3′-UTR of the HLA-G gene to comprehend their potential influence on gene expression and immune regulation. Polymorphisms in this region may impact the stability of HLA-G mRNA, thus affecting the levels of HLA-G protein. Numerous studies have examined the connection between specific 3′-UTR polymorphisms of HLA-G and the likelihood of experiencing RSA (Barbaro et al. 2023).
Our study demonstrated no significant difference between individual polymorphisms and RSA. Although there were differences between the case and control groups, they were not significant. However, analysis of the haplotypes between the two groups revealed significant differences in the two haplotypes. UTR-2 was more frequent in the group with RSA than in the healthy control group with statistical significance (p = 0.02). This haplotype represents a risk factor for RSA (OR = 1.78 and 95% CI = 1.12–2.84). In UTR-2, the 14 bp insertion polymorphism, as well as the +3003T, +3010C, +3142G, and +3196G alleles, contribute to the haplotype. Conversely, in the RSA group, UTR-4 had a lower frequency compared to the healthy group, signifying a statistically significant difference (p = 0.04). This finding highlights the protective role of the UTR-4 haplotype against RSA. The UTR-4 haplotype consists of polymorphisms, including the deletion of 14 bp, and +3003C, +3010G, +3142C, and +3196C alleles. Although there were differences in these polymorphisms alone between the two groups, they were not statistically significant. However, a significant difference was observed between the two groups regarding haplotypes.
The presence of a 14-bp sequence can serve as a hidden branch point for HLA-G mRNA splicing, which in some cases results in the elimination of a 92 nucleotide sequence. Although shorter mRNA sequences tend to be more stable, they have been found to correlate with reduced levels of the HLA-G protein. The insertion allele detected within the UTR-2 haplotype leads to diminished protein expression, potentially impacting maternal immune tolerance and contributing to RSA. On the other hand, the deletion allele found in the UTR-4 haplotype increases HLA-G protein expression, offering protection against abortion. Interestingly, this specific polymorphism alone does not exhibit any significant differences between the two groups under study. However, it may interact synergistically with other polymorphisms within the UTR-2 haplotype. Furthermore, another variant of this polymorphism involves a deletion of 14 base pairs, which is present in the protective UTR-4 haplotype and likely plays a crucial role in immune regulation (Castelli et al. 2009).
In UTR-2, there are C nucleotides at the +3010 site, which match the increased binding of some miRNAs, such as mir-513a, leading to a low level of HLA-g protein. On the other hand, the presence of G at +3010 in the UTR-4 haplotype causes a mismatch between microRNA and RNA, increases gene expression, and plays a protective role against abortion.
Our analysis aligns with previous studies that have highlighted the importance of specific SNPs in influencing HLA-G expression and their potential roles in pregnancy-related disorders. For instance, the presence of C nucleotides at the +3010 site in the UTR-2 haplotype correlates with increased binding of certain microRNAs, such as miR-513a, which can lead to reduced levels of HLA-G protein. In contrast, the G nucleotide at +3010 in the UTR-4 haplotype creates a mismatch with microRNA binding, thereby enhancing gene expression and providing a protective effect against miscarriage.
While our findings regarding UTR-2 and UTR-4 haplotypes contribute to the growing body of evidence linking HLA-G polymorphisms to reproductive outcomes, it is essential to note that other haplotypes did not show significant differences between cases and controls. This inconsistency emphasizes the complex genetic landscape surrounding RSA and preeclampsia (Nilsson et al. 2020).
Moreover, our results regarding the +3187A/G SNP are consistent with previous reports that have identified its association with various disorders, including preeclampsia. The G allele's higher frequency in our preeclampsia group compared to controls parallels findings from earlier studies that suggest this SNP may influence HLA-G expression through mechanisms involving mRNA stability (Quach et al. 2014).
It is essential to highlight that the HLA-G 3′-UTR haplotypes in this region are associated with distinct subsets of miRNAs that have the potential to bind and degrade the HLA-G mRNA. Interestingly, all 3′-UTR haplotypes containing the 14-bp sequence also possess the +3003T allele. Specific miRNAs with high affinity and others with low affinity target this allele. Furthermore, these haplotypes also exhibit the 3142G allele, which is strongly associated with multiple miRNAs with high-affinity binding (Castelli et al. 2009).
In addition to recurrent miscarriage, the HLA-G gene and its polymorphisms are associated with various pregnancy-related disorders, including preeclampsia. While the 14 bp insertion/deletion polymorphism did not show significant differences between cases and controls, its inconsistent associations with preeclampsia in previous studies suggest a complex genetic landscape. Notably, a significant correlation has been reported between the G/G genotype at +3187A/G and the C/C genotype at +3027A/C in placental tissues from patients with preeclampsia. The combination of G/G at +3187A/G and C/C at +3027A/C was found to be significantly more prevalent in preeclampsia cases, emphasizing the intricate nature of genetic interactions in multifactorial diseases like recurrent miscarriage and preeclampsia (Mandò et al. 2016; Quach et al. 2014).
This study had some limitations. Only 80 RSA women were evaluated, and it is suggested that more cases be assessed during future studies. Also, no anatomical, hormonal, or cytogenetic studies were performed.
In conclusion, this study analyzed the extended 3′-UTR HLA-G haplotypes for the first time in women with RSA in the northwestern population of Iran. In addition to investigating the association between each polymorphism in the 3′-UTR region of the HLA-G gene and RSA, this study has also analyzed and evaluated haplotypes within the 3′-UTR. This report represents the first comprehensive study conducted on the population of Northwestern Iran. Our results suggested no association between 3′UTR haplotypes of HLA-G and RSA. However, some alleles and genotypes were associated with RSA. This work supports the significance of HLA-G in pregnancy, even if a more thorough investigation that evaluates additional HLA-G gene segments, such as the promoter and the coding region, might help research HLA-G expression.
Author Contributions
A.M. experimented and wrote the manuscript. M.K.-K. supervised the work and contributed to the research design and implementation, the results analysis, and the manuscript's writing. M.H. helped supervise the project and contributed to data analysis. All authors discussed the results and contributed and reviewed the final manuscript.
Acknowledgments
We thank all patients and individuals for participating in this project.
Ethics Statement
This study was approved by the Medical Ethics Committee of the University of Tabriz (IR.TABRIZU.REC.1398.029).
Consent
All stages of sample collection have been done with the consent of patients and healthy people, and written consent forms have been obtained.
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.




