Directed Evolution of Piperazic Acid Incorporation by a Nonribosomal Peptide Synthetase**
Philipp Stephan
Junior Research Group Biosynthetic Design of Natural Products, Leibniz Institute for Natural Product Research and Infection Biology (HKI), Beutenbergstr. 11a, 07745 Jena, Germany
Search for more papers by this authorChloe Langley
Department of Natural Product Biosynthesis, Max Planck Institute for Chemical Ecology, Hans-Knöll-Str. 8, 07745 Jena, Germany
Search for more papers by this authorDaniela Winkler
Junior Research Group Biosynthetic Design of Natural Products, Leibniz Institute for Natural Product Research and Infection Biology (HKI), Beutenbergstr. 11a, 07745 Jena, Germany
Search for more papers by this authorDr. Jérôme Basquin
Department of Structural Cell Biology, Max Planck Institute for Biochemistry, Am Klopferspitz 18, 82152 Planegg Martinsried, Germany
Search for more papers by this authorDr. Lorenzo Caputi
Department of Natural Product Biosynthesis, Max Planck Institute for Chemical Ecology, Hans-Knöll-Str. 8, 07745 Jena, Germany
Search for more papers by this authorProf. Dr. Sarah E. O'Connor
Department of Natural Product Biosynthesis, Max Planck Institute for Chemical Ecology, Hans-Knöll-Str. 8, 07745 Jena, Germany
Search for more papers by this authorCorresponding Author
Dr. Hajo Kries
Junior Research Group Biosynthetic Design of Natural Products, Leibniz Institute for Natural Product Research and Infection Biology (HKI), Beutenbergstr. 11a, 07745 Jena, Germany
Search for more papers by this authorPhilipp Stephan
Junior Research Group Biosynthetic Design of Natural Products, Leibniz Institute for Natural Product Research and Infection Biology (HKI), Beutenbergstr. 11a, 07745 Jena, Germany
Search for more papers by this authorChloe Langley
Department of Natural Product Biosynthesis, Max Planck Institute for Chemical Ecology, Hans-Knöll-Str. 8, 07745 Jena, Germany
Search for more papers by this authorDaniela Winkler
Junior Research Group Biosynthetic Design of Natural Products, Leibniz Institute for Natural Product Research and Infection Biology (HKI), Beutenbergstr. 11a, 07745 Jena, Germany
Search for more papers by this authorDr. Jérôme Basquin
Department of Structural Cell Biology, Max Planck Institute for Biochemistry, Am Klopferspitz 18, 82152 Planegg Martinsried, Germany
Search for more papers by this authorDr. Lorenzo Caputi
Department of Natural Product Biosynthesis, Max Planck Institute for Chemical Ecology, Hans-Knöll-Str. 8, 07745 Jena, Germany
Search for more papers by this authorProf. Dr. Sarah E. O'Connor
Department of Natural Product Biosynthesis, Max Planck Institute for Chemical Ecology, Hans-Knöll-Str. 8, 07745 Jena, Germany
Search for more papers by this authorCorresponding Author
Dr. Hajo Kries
Junior Research Group Biosynthetic Design of Natural Products, Leibniz Institute for Natural Product Research and Infection Biology (HKI), Beutenbergstr. 11a, 07745 Jena, Germany
Search for more papers by this authorA previous version of this manuscript has been deposited on a preprint server (https://doi.org/10.1101/2023.04.03.535426).
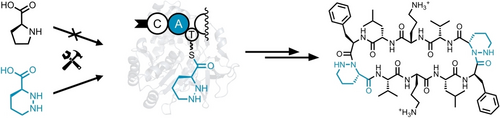
Graphical Abstract
Efficient directed evolution protocols for nonribosomal peptide synthetases are needed to adapt the structures of antibiotic peptides for the fight against antimicrobial resistance. Here, an easily reproducible directed evolution protocol was used to reprogram the synthetase for the antibiotic peptide gramicidin S. A few mutations were sufficient to incorporate the non-standard building block piperazic acid instead of proline with perfect specificity.
Abstract
Engineering of biosynthetic enzymes is increasingly employed to synthesize structural analogues of antibiotics. Of special interest are nonribosomal peptide synthetases (NRPSs) responsible for the production of important antimicrobial peptides. Here, directed evolution of an adenylation domain of a Pro-specific NRPS module completely switched substrate specificity to the non-standard amino acid piperazic acid (Piz) bearing a labile N−N bond. This success was achieved by UPLC-MS/MS-based screening of small, rationally designed mutant libraries and can presumably be replicated with a larger number of substrates and NRPS modules. The evolved NRPS produces a Piz-derived gramicidin S analogue. Thus, we give new impetus to the too-early dismissed idea that widely accessible low-throughput methods can switch the specificity of NRPSs in a biosynthetically useful fashion.
Open Research
Data Availability Statement
The coordinates and reflection data of the crystal structure described in this manuscript are available in the Protein Data Bank (PDB ID: 8P5O). Further primary data are shown in the Supporting Information and are available from the authors upon request.
Supporting Information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
| Filename | Description |
|---|---|
| anie202304843-sup-0001-misc_information.pdf4 MB | Supporting Information |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
References
- 1S. A. Sieber, M. A. Marahiel, Chem. Rev. 2005, 105, 715–738.
- 2R. D. Süssmuth, A. Mainz, Angew. Chem. Int. Ed. 2017, 56, 3770–3821.
- 3Y. Liu, S. Ding, J. Shen, K. Zhu, Nat. Prod. Rep. 2019, 36, 573–592.
- 4J. Krätzschmar, M. Krause, M. A. Marahiel, J. Bacteriol. 1989, 171, 5422–5429.
- 5M. Winn, J. K. Fyans, Y. Zhuo, J. Micklefield, Nat. Prod. Rep. 2016, 33, 317–347.
- 6A. S. Brown, M. J. Calcott, J. G. Owen, D. F. Ackerley, Nat. Prod. Rep. 2018, 35, 1210–1228.
- 7K. A. Bozhüyük, J. Micklefield, B. Wilkinson, Curr. Opin. Microbiol. 2019, 51, 88–96.
- 8C. Beck, J. F. G. Garzón, T. Weber, Biotechnol. Bioprocess Eng. 2020, 25, 886–894.
- 9M. J. Calcott, J. G. Owen, I. L. Lamont, D. F. Ackerley, Appl. Environ. Microbiol. 2014, 80, 5723–5731.
- 10H. Kries, D. L. Niquille, D. Hilvert, Chem. Biol. 2015, 22, 640–648.
- 11F. Yan, C. Burgard, A. Popoff, N. Zaburannyi, G. Zipf, J. Maier, H. S. Bernauer, S. C. Wenzel, R. Müller, Chem. Sci. 2018, 9, 7510–7519.
- 12K. A. J. Bozhüyük, F. Fleischhacker, A. Linck, F. Wesche, A. Tietze, C.-P. Niesert, H. B. Bode, Nat. Chem. 2018, 10, 275–281.
- 13M. Kaniusaite, R. J. A. Goode, J. Tailhades, R. B. Schittenhelm, M. J. Cryle, Chem. Sci. 2020, 11, 9443–9458.
- 14M. Schoppet, M. Peschke, A. Kirchberg, V. Wiebach, R. D. Süssmuth, E. Stegmann, M. J. Cryle, Chem. Sci. 2019, 10, 118–133.
- 15M. J. Calcott, J. G. Owen, D. F. Ackerley, Nat. Commun. 2020, 11, 4554.
- 16T. Izoré, Y. T. Candace Ho, J. A. Kaczmarski, A. Gavriilidou, K. H. Chow, D. L. Steer, R. J. A. Goode, R. B. Schittenhelm, J. Tailhades, M. Tosin, G. L. Challis, E. H. Krenske, N. Ziemert, C. J. Jackson, M. J. Cryle, Nat. Commun. 2021, 12, 2511.
- 17T. Stachelhaus, D. Mootz, A. Marahiel, Chem. Biol. 1999, 6, 493–505.
- 18G. L. Challis, J. Ravel, C. A. Townsend, Chem. Biol. 2000, 7, 211–224.
- 19F. Kudo, A. Miyanaga, T. Eguchi, J. Ind. Microbiol. Biotechnol. 2019, 46, 515–536.
- 20K. Eppelmann, T. Stachelhaus, M. A. Marahiel, Biochemistry 2002, 41, 9718–9726.
- 21J. Thirlway, R. Lewis, L. Nunns, M. Al Nakeeb, M. Styles, A. W. Struck, C. P. Smith, J. Micklefield, Angew. Chem. Int. Ed. 2012, 51, 7181–7184.
- 22J. W. Han, E. Y. Kim, J. M. Lee, Y. S. Kim, E. Bang, B. S. Kim, Biotechnol. Lett. 2012, 34, 1327–1334.
- 23H. Kries, R. Wachtel, A. Pabst, B. Wanner, D. Niquille, D. Hilvert, Angew. Chem. Int. Ed. 2014, 53, 10105–10108.
- 24H. Kaljunen, S. H. H. Schiefelbein, D. Stummer, S. Kozak, R. Meijers, G. Christiansen, A. Rentmeister, Angew. Chem. Int. Ed. 2015, 54, 8833–8836.
- 25M. Kaniusaite, T. Kittilä, R. J. A. Goode, R. B. Schittenhelm, M. J. Cryle, ACS Chem. Biol. 2020, 15, 2444–2455.
- 26M. A. Fischbach, J. R. Lai, E. D. Roche, C. T. Walsh, D. R. Liu, Proc. Natl. Acad. Sci. USA 2007, 104, 11951–11956.
- 27B. Villiers, F. Hollfelder, Chem. Biol. 2011, 18, 1290–1299.
- 28B. S. Evans, Y. Chen, W. W. Metcalf, H. Zhao, N. L. Kelleher, Chem. Biol. 2011, 18, 601–607.
- 29K. Zhang, K. M. Nelson, K. Bhuripanyo, K. D. Grimes, B. Zhao, C. C. Aldrich, J. Yin, Chem. Biol. 2013, 20, 92–101.
- 30D. L. Niquille, D. A. Hansen, T. Mori, D. Fercher, H. Kries, D. Hilvert, Nat. Chem. 2018, 10, 282–287.
- 31A. Camus, G. Truong, P. R. E. Mittl, G. Markert, D. Hilvert, J. Am. Chem. Soc. 2022, 144, 17567–17575.
- 32A. J. Oelke, D. J. France, T. Hofmann, G. Wuitschik, S. V. Ley, Nat. Prod. Rep. 2011, 28, 1445–1471.
- 33K. D. Morgan, R. J. Andersen, K. S. Ryan, Nat. Prod. Rep. 2019, 36, 1628–1653.
- 34Z.-W. Wei, H. Niikura, K. D. Morgan, C. M. Vacariu, R. J. Andersen, K. S. Ryan, J. Am. Chem. Soc. 2022, 144, 13556–13564.
- 35N. Xi, L. B. Alemany, M. A. Ciufolini, J. Am. Chem. Soc. 1998, 120, 80–86.
- 36C. S. Neumann, W. Jiang, J. R. Heemstra, E. A. Gontang, R. Kolter, C. T. Walsh, ChemBioChem 2012, 13, 972–976.
- 37Y. L. Du, H. Y. He, M. A. Higgins, K. S. Ryan, Nat. Chem. Biol. 2017, 13, 836–838.
- 38A. J. Waldman, T. L. Ng, P. Wang, E. P. Balskus, Chem. Rev. 2017, 117, 5784–5863.
- 39M. Tamaki, T. Harada, K. Fujinuma, K. Takanashi, M. Shindo, M. Kimura, Y. Uchida, Chem. Pharm. Bull. 2012, 60, 1134–1138.
- 40H. Zhao, L. Wang, D. Wan, J. Qi, R. Gong, Z. Deng, W. Chen, Microb. Cell Fact. 2016, 15, 160–172.
- 41Y. Du, Y. Wang, T. Huang, M. Tao, Z. Deng, S. Lin, BMC Microbiol. 2014, 14, 1471–2180.
- 42D. G. Fujimori, S. Hrvatin, C. S. Neumann, M. Strieker, M. A. Marahiel, C. T. Walsh, Proc. Natl. Acad. Sci. USA 2007, 104, 16498–16503.
- 43J. Ma, Z. Wang, H. Huang, M. Luo, D. Zuo, B. Wang, A. Sun, Y.-Q. Cheng, C. Zhang, J. Ju, Angew. Chem. Int. Ed. 2011, 50, 7797–7802.
- 44M. Röttig, M. H. Medema, K. Blin, T. Weber, C. Rausch, O. Kohlbacher, Nucleic Acids Res. 2011, 39, 362–367.
- 45G. Qu, A. Li, C. G. Acevedo-Rocha, Z. Sun, M. T. Reetz, Angew. Chem. Int. Ed. 2020, 59, 13204–13231.
- 46A. Miyanaga, J. Cieślak, Y. Shinohara, F. Kudo, T. Eguchi, J. Biol. Chem. 2014, 289, 31448–31457.
- 47E. Conti, T. Stachelhaus, M. a. Marahiel, P. Brick, EMBO J. 1997, 16, 4174–4183.
- 48M. C. Lux, L. C. Standke, D. S. Tan, J. Antibiot. 2019, 72, 325–349.
- 49D. J. Wilson, C. C. Aldrich, Anal. Biochem. 2010, 404, 56–63.
- 50E. J. Drake, B. R. Miller, C. Shi, J. T. Tarrasch, J. A. Sundlov, C. Leigh Allen, G. Skiniotis, C. C. Aldrich, A. M. Gulick, Nature 2016, 529, 235–238.
- 51A. Stanišić, A. Hüsken, P. Stephan, D. L. Niquille, J. Reinstein, H. Kries, ACS Catal. 2021, 11, 8692–8700.
- 52I. Folger, N. Frota, A. Pistofidis, D. Niquille, D. Hansen, T. M. Schmeing, D. Hilvert, Research Square preprint 2023, https://doi.org/10.21203/rs.3.rs-2531419/v1.
10.21203/rs.3.rs-2531419/v1 Google Scholar
- 53F. Pourmasoumi, S. Hengoju, K. Beck, P. Stephan, L. Klopfleisch, M. Hoernke, M. A. Rosenbaum, H. Kries, BioRxiv preprint 2023, https://doi.org/10.1101/2023.01.13.523969.
10.1101/2023.01.13.523969 Google Scholar
- 54M. S. Packer, D. R. Liu, Nat. Rev. Genet. 2015, 16, 379–394.
- 55F. H. Arnold, Angew. Chem. Int. Ed. 2018, 57, 4143–4148.
- 56Y. Wang, P. Xue, M. Cao, T. Yu, S. T. Lane, H. Zhao, Chem. Rev. 2021, 121, 12384–12444.
- 57M. A. Ciufolini, N. Xi, Chem. Soc. Rev. 1998, 27, 437–445.
- 58M. A. Ciufolini, T. Shimizu, S. Swaminathan, N. Xi, Tetrahedron Lett. 1997, 38, 4947–4950.