A DNA Barcode-Based Aptasensor Enables Rapid Testing of Porcine Epidemic Diarrhea Viruses in Swine Saliva Using Electrochemical Readout
Amanda Victorious
School of Biomedical Engineering, McMaster University, 1280 Main Street West, Hamilton, Ontario, L8S 4K1 Canada
These authors contributed equally to this work.
Search for more papers by this authorDr. Zijie Zhang
Department of Biochemistry and Biomedical Sciences, McMaster University, Canada
These authors contributed equally to this work.
Search for more papers by this authorDr. Dingran Chang
Department of Biochemistry and Biomedical Sciences, McMaster University, Canada
Search for more papers by this authorRoderick Maclachlan
Department of Engineering Physics, McMaster University, Canada
Search for more papers by this authorDr. Richa Pandey
School of Biomedical Engineering, McMaster University, 1280 Main Street West, Hamilton, Ontario, L8S 4K1 Canada
Search for more papers by this authorJianrun Xia
Department of Biochemistry and Biomedical Sciences, McMaster University, Canada
Search for more papers by this authorJimmy Gu
Department of Biochemistry and Biomedical Sciences, McMaster University, Canada
Search for more papers by this authorProf. Dr. Todd Hoare
School of Biomedical Engineering, McMaster University, 1280 Main Street West, Hamilton, Ontario, L8S 4K1 Canada
Department of Chemical Engineering, McMaster University, Canada
Search for more papers by this authorCorresponding Author
Prof. Dr. Leyla Soleymani
School of Biomedical Engineering, McMaster University, 1280 Main Street West, Hamilton, Ontario, L8S 4K1 Canada
Department of Engineering Physics, McMaster University, Canada
Michael G. DeGroote Institute for Infectious Disease Research, McMaster University, Canada
Search for more papers by this authorCorresponding Author
Prof. Dr. Yingfu Li
School of Biomedical Engineering, McMaster University, 1280 Main Street West, Hamilton, Ontario, L8S 4K1 Canada
Department of Biochemistry and Biomedical Sciences, McMaster University, Canada
Michael G. DeGroote Institute for Infectious Disease Research, McMaster University, Canada
Search for more papers by this authorAmanda Victorious
School of Biomedical Engineering, McMaster University, 1280 Main Street West, Hamilton, Ontario, L8S 4K1 Canada
These authors contributed equally to this work.
Search for more papers by this authorDr. Zijie Zhang
Department of Biochemistry and Biomedical Sciences, McMaster University, Canada
These authors contributed equally to this work.
Search for more papers by this authorDr. Dingran Chang
Department of Biochemistry and Biomedical Sciences, McMaster University, Canada
Search for more papers by this authorRoderick Maclachlan
Department of Engineering Physics, McMaster University, Canada
Search for more papers by this authorDr. Richa Pandey
School of Biomedical Engineering, McMaster University, 1280 Main Street West, Hamilton, Ontario, L8S 4K1 Canada
Search for more papers by this authorJianrun Xia
Department of Biochemistry and Biomedical Sciences, McMaster University, Canada
Search for more papers by this authorJimmy Gu
Department of Biochemistry and Biomedical Sciences, McMaster University, Canada
Search for more papers by this authorProf. Dr. Todd Hoare
School of Biomedical Engineering, McMaster University, 1280 Main Street West, Hamilton, Ontario, L8S 4K1 Canada
Department of Chemical Engineering, McMaster University, Canada
Search for more papers by this authorCorresponding Author
Prof. Dr. Leyla Soleymani
School of Biomedical Engineering, McMaster University, 1280 Main Street West, Hamilton, Ontario, L8S 4K1 Canada
Department of Engineering Physics, McMaster University, Canada
Michael G. DeGroote Institute for Infectious Disease Research, McMaster University, Canada
Search for more papers by this authorCorresponding Author
Prof. Dr. Yingfu Li
School of Biomedical Engineering, McMaster University, 1280 Main Street West, Hamilton, Ontario, L8S 4K1 Canada
Department of Biochemistry and Biomedical Sciences, McMaster University, Canada
Michael G. DeGroote Institute for Infectious Disease Research, McMaster University, Canada
Search for more papers by this authorGraphical Abstract
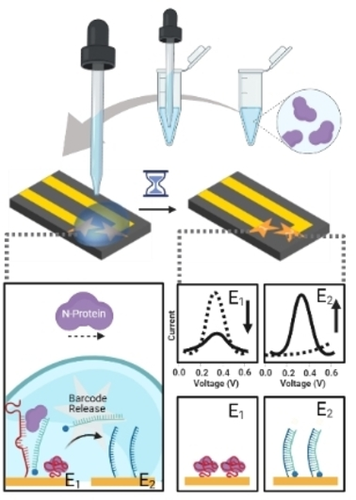
A rapid, simple, and reagent-less dual-electrode electrochemical chip (DEE-Chip) was developed using a barcode-releasing electroactive aptamer for on-farm detection of porcine epidemic diarrhea viruses (PEDv). This biosensor demonstrated a diagnostic sensitivity of 83 % and specificity of 100 % using 12 porcine saliva samples with a concordance value of 92 % at an analysis time of one hour.
Abstract
Pen-side testing of farm animals for infectious diseases is critical for preventing transmission in herds and providing timely intervention. However, most existing pathogen tests have to be conducted in centralized labs with sample-to-result times of 2–4 days. Herein we introduce a test that uses a dual-electrode electrochemical chip (DEE-Chip) and a barcode-releasing electroactive aptamer for rapid on-farm detection of porcine epidemic diarrhea viruses (PEDv). The sensor exploits inter-electrode spacing reduction and active field mediated transport to accelerate barcode movement from electroactive aptamers to the detection electrode, thus expediting assay operation. The test yielded a clinically relevant limit-of-detection of 6 nM (0.37 μg mL−1) in saliva-spiked PEDv samples. Clinical evaluation of this biosensor with 12 porcine saliva samples demonstrated a diagnostic sensitivity of 83 % and specificity of 100 % with a concordance value of 92 % at an analysis time of one hour.
Conflict of interest
The authors declare no conflict of interest.
Supporting Information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
| Filename | Description |
|---|---|
| anie202204252-sup-0001-misc_information.pdf1.3 MB | Supporting Information |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
References
- 1J. Cui, F. Li, Z.-L. Shi, Nat. Rev. Microbiol. 2019, 17, 181–192.
- 2S. N. Bevins, M. Lutman, K. Pedersen, N. Barrett, T. Gidlewski, T. J. Deliberto, A. B. Franklin, Emerging Infect. Dis. 2018, 24, 1390–1392.
- 3L. L. Schulz, G. T. Tonsor, J. Anim. Sci. 2015, 93, 5111–5118.
- 4A. Bang, S. Khadakkar, Proc. Natl. Acad. Sci. USA 2020, 117, 29995–29999.
- 5S. Chen, L. Zhang, L. Wang, H. Ouyang, L. Ren, Proc. Natl. Acad. Sci. USA 2021, 118, e2022344118.
- 6J. Vidic, M. Manzano, C.-M. Chang, N. Jaffrezic-Renault, Vet. Res. 2017, 48, 11.
- 7A. Cullinane, M. Garvey, Rev. Sci. Tech. 2021, 40, 75–89.
- 8S. Neethirajan, S. K. Tuteja, S.-T. Huang, D. Kelton, Biosens. Bioelectron. 2017, 98, 398–407.
- 9A. Kang, M. Yeom, H. Kim, S.-W. Yoon, D.-G. Jeong, H.-J. Moon, K.-S. Lyoo, W. Na, D. Song, Immune Netw. 2021, 21, e11.
- 10S. Zou, L. Wu, G. Li, J. Wang, D. Cao, T. Xu, A. Jia, Y. Tang, Anim. Dis. 2021, 1, 27.
- 11W. Xiao, C. Huang, F. Xu, J. Yan, H. Bian, Q. Fu, K. Xie, L. Wang, Y. Tang, Sens. Actuators B 2018, 266, 63–70.
- 12B. Fan, J. Sun, L. Zhu, J. Zhou, Y. Zhao, Z. Yu, B. Sun, R. Guo, K. He, B. Li, Front. Vet. Sci. 2020, 7, 540248.
- 13K. Poonsuk, T.-Y. Cheng, J. Ji, J. Zimmerman, L. Giménez-Lirola, Porc. Health Manag. 2018, 4, 31.
- 14I. Díaz, J. Pujols, E. Cano, R. Cuadrado, N. Navarro, E. Mateu, M. Martín, Vet. Immunol. Immunopathol. 2021, 234, 110206.
- 15J. S. Oh, D. S. Song, J. S. Yang, J. Y. Song, H. J. Moon, T. Y. Kim, B. K. Park, J. Vet. Sci. 2005, 6, 349–352.
- 16Z. Ma, T. Wang, Z. Li, X. Guo, Y. Tian, Y. Li, S. Xiao, J. Nanobiotechnol. 2019, 17, 96.
- 17W.-T. Hsu, C.-Y. Chang, C.-H. Tsai, S.-C. Wei, H.-R. Lo, R. J. S. Lamis, H.-W. Chang, Y.-C. Chao, Viruses 2021, 13, 303.
- 18J. Ma, L. Wu, Z. Li, Z. Lu, W. Yin, A. Nie, F. Ding, B. Wang, H. Han, Anal. Chem. 2018, 90, 7415–7421.
- 19J. Ma, W. Wang, Y. Li, Z. Lu, X. Tan, H. Han, Anal. Chem. 2021, 93, 2090–2096.
- 20X. Li, Y. Wang, X. Zhang, Y. Gao, C. Sun, Y. Ding, F. Feng, W. Jin, G. Yang, Microchim. Acta 2020, 187, 217.
- 21E. M. McConnell, I. Cozma, D. Morrison, Y. Li, Anal. Chem. 2020, 92, 327–344.
- 22M. N. Kammer, A. Kussrow, I. Gandhi, R. Drabek, R. H. Batchelor, G. W. Jackson, D. J. Bornhop, Anal. Chem. 2019, 91, 10582–10588.
- 23J. S. Swensen, Y. Xiao, B. S. Ferguson, A. A. Lubin, R. Y. Lai, A. J. Heeger, K. W. Plaxco, H. Tom Soh, J. Am. Chem. Soc. 2009, 131, 4262–4266.
- 24Z. Zhang, R. Pandey, J. Li, J. Gu, D. White, H. D. Stacey, J. C. Ang, C. Steinberg, A. Capretta, C. D. M. Filipe, K. Mossman, C. Balion, M. S. Miller, B. J. Salena, D. Yamamura, L. Soleymani, J. D. Brennan, Y. Li, Angew. Chem. Int. Ed. 2021, 60, 24266–24274; Angew. Chem. 2021, 133, 24468–24476.
- 25Z. Zhang, J. Li, J. Gu, R. Amini, H. D. Stacey, J. C. Ang, D. White, C. D. M. Filipe, K. Mossman, M. S. Miller, B. J. Salena, D. Yamamura, P. Sen, L. Soleymani, J. D. Brennan, Y. Li, Chem. Eur. J. 2022, 28, e202200078.
- 26J. Li, Z. Zhang, J. Gu, H. D. Stacey, J. C. Ang, A. Capretta, C. D. M. Filipe, K. L. Mossman, C. Balion, B. J. Salena, D. Yamamura, L. Soleymani, M. S. Miller, J. D. Brennan, Y. Li, Nucleic Acids Res. 2021, 49, 7267–7279.
- 27D. Song, B. Park, Virus Genes 2012, 44, 167–175.
- 28T. A. Feagin, N. Maganzini, H. T. Soh, ACS Sens. 2018, 3, 1611–1615.
- 29L. R. Schoukroun-Barnes, F. C. Macazo, B. Gutierrez, J. Lottermoser, J. Liu, R. J. White, Annu. Rev. Anal. Chem. 2016, 9, 163–181.
- 30R. J. White, A. A. Rowe, K. W. Plaxco, Analyst 2010, 135, 589–594.
- 31Y. Xiao, T. Uzawa, R. J. White, D. DeMartini, K. W. Plaxco, Electroanalysis 2009, 21, 1267–1271.
- 32Y. Xiao, A. A. Lubin, A. J. Heeger, K. W. Plaxco, Angew. Chem. Int. Ed. 2005, 44, 5456–5459; Angew. Chem. 2005, 117, 5592–5595.
- 33D.-Y. Liu, Y. Zhao, X.-W. He, X.-B. Yin, Biosens. Bioelectron. 2011, 26, 2905–2910.
- 34Z. Tang, P. Mallikaratchy, R. Yang, Y. Kim, Z. Zhu, H. Wang, W. Tan, J. Am. Chem. Soc. 2008, 130, 11268–11269.
- 35Y. Xiao, B. D. Piorek, K. W. Plaxco, A. J. Heeger, J. Am. Chem. Soc. 2005, 127, 17990–17991.
- 36R. Liu, Z. Yang, Q. Guo, J. Zhao, J. Ma, Q. Kang, Y. Tang, Y. Xue, X. Lou, M. He, Electrochim. Acta 2015, 182, 516–523.
- 37K. Feng, C. Sun, Y. Kang, J. Chen, J.-H. Jiang, G.-L. Shen, R.-Q. Yu, Electrochem. Commun. 2008, 10, 531–535.
- 38Y. Xiao, X. Qu, K. W. Plaxco, A. J. Heeger, J. Am. Chem. Soc. 2007, 129, 11896–11897.
- 39M. Liu, J. Wang, Y. Chang, Q. Zhang, D. Chang, C. Y. Hui, J. D. Brennan, Y. Li, Angew. Chem. Int. Ed. 2020, 59, 7706–7710; Angew. Chem. 2020, 132, 7780–7784.
- 40B. Liwnaree, J. Narkpuk, S. Sungsuwan, A. Jongkaewwattana, P. Jaru-Ampornpan, PLoS One 2019, 14, e0212632.
- 41F. Deng, G. Ye, Q. Liu, M. T. Navid, X. Zhong, Y. Li, C. Wan, S. Xiao, Q. He, Z. F. Fu, G. Peng, Viruses 2016, 8, 55.
- 42R. Gysbers, K. Tram, J. Gu, Y. Li, Sci. Rep. 2015, 5, 11405.
- 43D. Chang, S. Zakaria, S. Esmaeili Samani, Y. Chang, C. D. Filipe, L. Soleymani, J. D. Brennan, M. Liu, Y. Li, Acc. Chem. Res. 2021, 54, 3540–3549.
- 44M. Elbadawi, J. J. Ong, T. D. Pollard, S. Gaisford, A. W. Basit, Adv. Funct. Mater. 2021, 31, 2006407.
- 45I. Russo Krauss, A. Merlino, A. Randazzo, E. Novellino, L. Mazzarella, F. Sica, Nucleic Acids Res. 2012, 40, 8119–8128.
- 46L. Soleymani, Z. Fang, E. H. Sargent, S. O. Kelley, Nat. Nanotechnol. 2009, 4, 844–848.
- 47L. Soleymani, Z. Fang, X. Sun, H. Yang, B. J. Taft, E. H. Sargent, S. O. Kelley, Angew. Chem. Int. Ed. 2009, 48, 8457–8460; Angew. Chem. 2009, 121, 8609–8612.
- 48G. Rabai, M. Orban, I. R. Epstein, Acc. Chem. Res. 1990, 23, 258–263.
- 49S. M. Traynor, G. A. Wang, R. Pandey, F. Li, L. Soleymani, Angew. Chem. Int. Ed. 2020, 59, 22617–22622; Angew. Chem. 2020, 132, 22806–22811.
- 50R. Pandey, Y. Lu, E. Osman, S. Saxena, Z. Zhang, S. Qian, A. Pollinzi, M. Smieja, Y. Li, L. Soleymani, T. Hoare, ACS Sens. 2022, 7, 985–994.
- 51F. Kuralay, S. Campuzano, J. Wang, Talanta 2012, 99, 155–160.
- 52P. Wu, D. W. Grainger, J. Proteome Res. 2006, 5, 2956–2965.
- 53Z. Chen, Q. Wu, J. Chen, X. Ni, J. Dai, Virol. Sin. 2020, 35, 351–354.
- 54R. Wang, J. Zhao, T. Jiang, Y. M. Kwon, H. Lu, P. Jiao, M. Liao, Y. Li, J. Virol. Methods 2013, 189, 362–369.
- 55K. Percze, Z. Szakács, É. Scholz, J. András, Z. Szeitner, C. H. Kieboom, G. Ferwerda, M. I. de Jonge, R. E. Gyurcsányi, T. Mészáros, Sci. Rep. 2017, 7, 42794.
- 56X. Zou, J. Wu, J. Gu, L. Shen, L. Mao, Front. Microbiol. 2019, 10, 1462.
- 57S. Dolai, M. Tabib-Azar, Med. Devices Sens. 2020, 3, e10112.
- 58J. Kang, G. Yeom, S.-J. Ha, M.-G. Kim, New J. Chem. 2019, 43, 6883–6889.
- 59S. O. Kelley, J. K. Barton, N. M. Jackson, M. G. Hill, Bioconjugate Chem. 1997, 8, 31–37.
- 60K. Ngamchuea, C. Batchelor-McAuley, R. G. Compton, Sens. Actuators B 2018, 262, 404–410.
- 61K. L. Baker, P. R. Thomas, L. A. Karriker, A. Ramirez, J. Zhang, C. Wang, D. J. Holtkamp, BMC Vet. Res. 2017, 13, 372.
- 62J. Bjustrom-Kraft, K. Woodard, L. Giménez-Lirola, M. Rotolo, C. Wang, Y. Sun, P. Lasley, J. Zhang, D. Baum, P. Gauger, BMC Vet. Res. 2016, 12, 99.
- 63R. Pandey, D. Chang, M. Smieja, T. Hoare, Y. Li, L. Soleymani, Nat. Chem. 2021, 13, 895–901.
- 64G. C. Mak, P. K. Cheng, S. S. Lau, K. K. Wong, C. Lau, E. T. Lam, R. C. Chan, D. N. Tsang, J. Clin. Virol. 2020, 129, 104500.
- 65L. Soleymani, F. Li, ACS Sens. 2017, 2, 458–467.
- 66A. Werner, Nucleic Acids Res. 2011, 39, e17–e17.
- 67M. H. Zweig, G. Campbell, Clin. Chem. 1993, 39, 561–577.
- 68J. N. Mandrekar, J. Thorac. Oncol. 2010, 5, 1315–1316.
- 69World Health Organization, Antigen-Detection in the Diagnosis of SARS-CoV-2 Infection: Interim Guidance, 6 October 2021, World Health Organization, 2021.