CRISPR Mediated Biosensing Toward Understanding Cellular Biology and Point-of-Care Diagnosis
Corresponding Author
Dr. Yifan Dai
Department of Biomedical Engineering, Duke University, Durham, North Carolina, 27708 USA
Department of Chemical and Biomolecular Engineering, Case Western Reserve University, Cleveland, Ohio, 44106 USA
These authors contributed equally to this work.
Search for more papers by this authorCorresponding Author
Dr. Yanfang Wu
School of Chemistry, Australian Centre for NanoMedicine, and ARC Centre of Excellence in Convergent Bio-Nano Science and Technology, The University of New South Wales, Sydney, NSW, 2052 Australia
These authors contributed equally to this work.
Search for more papers by this authorProf. Guozhen Liu
Graduate School of Biomedical Engineering, The University of New South Wales, Sydney, NSW, 2052 Australia
Search for more papers by this authorProf. J. Justin Gooding
School of Chemistry, Australian Centre for NanoMedicine, and ARC Centre of Excellence in Convergent Bio-Nano Science and Technology, The University of New South Wales, Sydney, NSW, 2052 Australia
Search for more papers by this authorCorresponding Author
Dr. Yifan Dai
Department of Biomedical Engineering, Duke University, Durham, North Carolina, 27708 USA
Department of Chemical and Biomolecular Engineering, Case Western Reserve University, Cleveland, Ohio, 44106 USA
These authors contributed equally to this work.
Search for more papers by this authorCorresponding Author
Dr. Yanfang Wu
School of Chemistry, Australian Centre for NanoMedicine, and ARC Centre of Excellence in Convergent Bio-Nano Science and Technology, The University of New South Wales, Sydney, NSW, 2052 Australia
These authors contributed equally to this work.
Search for more papers by this authorProf. Guozhen Liu
Graduate School of Biomedical Engineering, The University of New South Wales, Sydney, NSW, 2052 Australia
Search for more papers by this authorProf. J. Justin Gooding
School of Chemistry, Australian Centre for NanoMedicine, and ARC Centre of Excellence in Convergent Bio-Nano Science and Technology, The University of New South Wales, Sydney, NSW, 2052 Australia
Search for more papers by this authorGraphical Abstract
Abstract
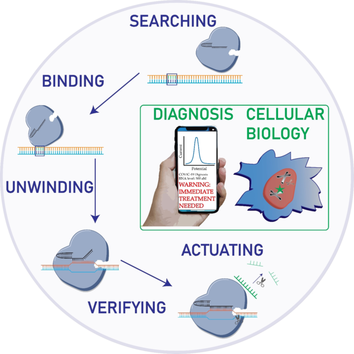
Recent advances in CRISPR based biotechnologies have greatly expanded our capabilities to repurpose CRISPR for the development of biomolecular sensors for diagnosing diseases and understanding cellular pathways. The key attribute that allows CRISPR to be widely utilized is the programmable and highly selective mechanism. In this Minireview, we first illustrate the molecular principle of CRISPR functioning process from sensing to actuating. Next, the CRISPR based biosensing strategies for nucleic acids, proteins and small molecules are summarized. We highlight some of recent advances in applications for in vitro detection of biomolecules and in vivo imaging of cellular networks. Finally, the challenges with, and exciting prospects of, CRISPR based biosensing developments are discussed.
Conflict of interest
The authors declare no conflict of interest.
References
- 1P. C. Ng, S. S. Murray, S. Levy, J. C. Venter, Nature 2009, 461, 724–726.
- 2
- 2aK. M. Esvelt, P. Mali, J. L. Braff, M. Moosburner, S. J. Yaung, G. M. Church, Nat. Methods 2013, 10, 1116;
- 2bY. E. Tak, B. P. Kleinstiver, J. K. Nuñez, J. Y. Hsu, J. E. Horng, J. Gong, J. S. Weissman, J. K. Joung, Nat. Methods 2017, 14, 1163–1166;
- 2cJ. S. Gootenberg, O. O. Abudayyeh, J. W. Lee, P. Essletzbichler, A. J. Dy, J. Joung, V. Verdine, N. Donghia, N. M. Daringer, C. A. Freije, Science 2017, 356, 438–442.
- 3Y. Wu, R. D. Tilley, J. J. Gooding, J. Am. Chem. Soc. 2019, 141, 1162–1170.
- 4Y. Dai, C. C. Liu, Angew. Chem. Int. Ed. 2019, 58, 12355–12368; Angew. Chem. 2019, 131, 12483–12496.
- 5R. Barrangou, C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero, P. Horvath, Science 2007, 315, 1709–1712.
- 6E. V. Koonin, K. S. Makarova, F. Zhang, Curr. Opin. Microbiol. 2017, 37, 67–78.
- 7K. S. Makarova, F. Zhang, E. V. Koonin, Cell 2017, 168, 946–946.e1.
- 8S. Shmakov, O. O. Abudayyeh, K. S. Makarova, Y. I. Wolf, J. S. Gootenberg, E. Semenova, L. Minakhin, J. Joung, S. Konermann, K. Severinov, Mol. Cell 2015, 60, 385–397.
- 9
- 9aG. J. Knott, J. A. Doudna, Science 2018, 361, 866–869;
- 9bP. D. Hsu, E. S. Lander, F. Zhang, Cell 2014, 157, 1262–1278;
- 9cP. Mali, L. Yang, K. M. Esvelt, J. Aach, M. Guell, J. E. DiCarlo, J. E. Norville, G. M. Church, Science 2013, 339, 823–826;
- 9dB. Zetsche, J. S. Gootenberg, O. O. Abudayyeh, I. M. Slaymaker, K. S. Makarova, P. Essletzbichler, S. E. Volz, J. Joung, J. van der Oost, A. Regev, E. V. Koonin, F. Zhang, Cell 2015, 163, 759–771.
- 10
- 10aF. A. Ran, P. D. Hsu, J. Wright, V. Agarwala, D. A. Scott, F. Zhang, Nat. Protoc. 2013, 8, 2281–2308;
- 10bJ. S. Chen, J. A. Doudna, Nat. Rev. Chem. 2017, 1, 0078.
- 11C. Anders, O. Niewoehner, A. Duerst, M. Jinek, Nature 2014, 513, 569–573.
- 12
- 12aM. M. Jore, M. Lundgren, E. van Duijn, J. B. Bultema, E. R. Westra, S. P. Waghmare, B. Wiedenheft, Ü. Pul, R. Wurm, R. Wagner, M. R. Beijer, A. Barendregt, K. Zhou, A. P. L. Snijders, M. J. Dickman, J. A. Doudna, E. J. Boekema, A. J. R. Heck, J. van der Oost, S. J. J. Brouns, Nat. Struct. Mol. Biol. 2011, 18, 529–536;
- 12bD. C. Swarts, J. van der Oost, M. Jinek, Mol. Cell 2017, 66, 221–233.
- 13L. A. Marraffini, E. J. Sontheimer, Nature 2010, 463, 568–571.
- 14
- 14aJ. S. Chen, E. Ma, L. B. Harrington, M. Da Costa, X. Tian, J. M. Palefsky, J. A. Doudna, Science 2018, 360, 436–439;
- 14bJ. S. Gootenberg, O. O. Abudayyeh, M. J. Kellner, J. Joung, J. J. Collins, F. Zhang, Science 2018, 360, 439–444.
- 15A.-C. Gourinat, O. O'Connor, E. Calvez, C. Goarant, M. Dupont-Rouzeyrol, Emerging Infect. Dis. 2015, 21, 84–86.
- 16K. Pardee, A. A. Green, M. K. Takahashi, D. Braff, G. Lambert, J. W. Lee, T. Ferrante, D. Ma, N. Donghia, M. Fan, N. M. Daringer, I. Bosch, D. M. Dudley, D. H. O'Connor, L. Gehrke, J. J. Collins, Cell 2016, 165, 1255–1266.
- 17A. A. Green, P. A. Silver, J. J. Collins, P. Yin, Cell 2014, 159, 925–939.
- 18W. Zhou, L. Hu, L. Ying, Z. Zhao, P. K. Chu, X.-F. Yu, Nat. Commun. 2018, 9, 5012.
- 19L. A. Gilbert, M. H. Larson, L. Morsut, Z. Liu, G. A. Brar, S. E. Torres, N. Stern-Ginossar, O. Brandman, E. H. Whitehead, J. A. Doudna, Cell 2013, 154, 442–451.
- 20G. T. Hess, L. Frésard, K. Han, C. H. Lee, A. Li, K. A. Cimprich, S. B. Montgomery, M. C. Bassik, Nat. Methods 2016, 13, 1036–1042.
- 21B. Chen, L. A. Gilbert, B. A. Cimini, J. Schnitzbauer, W. Zhang, G.-W. Li, J. Park, E. H. Blackburn, J. S. Weissman, L. S. Qi, B. Huang, Cell 2013, 155, 1479–1491.
- 22
- 22aJ. Y. Kishi, S. W. Lapan, B. J. Beliveau, E. R. West, A. Zhu, H. M. Sasaki, S. K. Saka, Y. Wang, C. L. Cepko, P. Yin, Nat. Methods 2019, 16, 533–544;
- 22bG. Wallner, R. Amann, W. Beisker, Cytometry 1993, 14, 136–143.
- 23
- 23aD. Samanta, S. B. Ebrahimi, C. A. Mirkin, Adv. Mater. 2020, 32, 1901743;
- 23bZ. Qing, J. Xu, J. Hu, J. Zheng, L. He, Z. Zou, S. Yang, W. Tan, R. Yang, Angew. Chem. Int. Ed. 2019, 58, 11574–11585; Angew. Chem. 2019, 131, 11698–11709;
- 23cF. Yang, Q. Li, L. Wang, G.-J. Zhang, C. Fan, ACS Sens. 2018, 3, 903–919.
- 24H. Wang, M. Nakamura, T. R. Abbott, D. Zhao, K. Luo, C. Yu, C. M. Nguyen, A. Lo, T. P. Daley, M. La Russa, Y. Liu, L. S. Qi, Science 2019, 365, 1301–1305.
- 25T. Weber, B. Wefers, W. Wurst, S. Sander, K. Rajewsky, R. Kühn, Nat. Biotechnol. 2015, 33, 543–548.
- 26Y. Zhang, L. Qian, W. Wei, Y. Wang, B. Wang, P. Lin, W. Liu, L. Xu, X. Li, D. Liu, S. Cheng, J. Li, Y. Ye, H. Li, X. Zhang, Y. Dong, X. Zhao, C. Liu, H. M. Zhang, Q. Ouyang, C. Lou, ACS Synth. Biol. 2017, 6, 211–216.
- 27S. S. Shekhawat, I. Ghosh, Curr. Opin. Chem. Biol. 2011, 15, 789–797.
- 28S. M. Marques, J. C. Esteves da Silva, IUBMB life 2009, 61, 6–17.
- 29D. A. Nelles, M. Y. Fang, M. R. O'Connell, J. L. Xu, S. J. Markmiller, J. A. Doudna, G. W. Yeo, Cell 2016, 165, 488–496.
- 30S.-Y. Li, Q.-X. Cheng, J.-K. Liu, X.-Q. Nie, G.-P. Zhao, J. Wang, Cell Res. 2018, 28, 491–493.
- 31D. C. Swarts, M. Jinek, Mol. Cell 2019, 73, 589–600.
- 32L. Liu, X. Li, J. Wang, M. Wang, P. Chen, M. Yin, J. Li, G. Sheng, Y. Wang, Cell 2017, 168, 121–134.
- 33S.-Y. Li, Q.-X. Cheng, J.-M. Wang, X.-Y. Li, Z.-L. Zhang, S. Gao, R.-B. Cao, G.-P. Zhao, J. Wang, Cell Discovery 2018, 4, 20.
- 34Y. Dai, R. A. Somoza, L. Wang, J. F. Welter, Y. Li, A. I. Caplan, C. C. Liu, Angew. Chem. Int. Ed. 2019, 58, 17399–17405; Angew. Chem. 2019, 131, 17560–17566.
- 35
- 35aB. P. Kleinstiver, M. S. Prew, S. Q. Tsai, V. V. Topkar, N. T. Nguyen, Z. Zheng, A. P. W. Gonzales, Z. Li, R. T. Peterson, J.-R. J. Yeh, M. J. Aryee, J. K. Joung, Nature 2015, 523, 481–485;
- 35bL. Gao, D. B. T. Cox, W. X. Yan, J. C. Manteiga, M. W. Schneider, T. Yamano, H. Nishimasu, O. Nureki, N. Crosetto, F. Zhang, Nat. Biotechnol. 2017, 35, 789–792.
- 36P. Qin, M. Park, K. J. Alfson, M. Tamhankar, R. Carrion, J. L. Patterson, A. Griffiths, Q. He, A. Yildiz, R. Mathies, K. Du, ACS Sens. 2019, 4, 1048–1054.
- 37C. Myhrvold, C. A. Freije, J. S. Gootenberg, O. O. Abudayyeh, H. C. Metsky, A. F. Durbin, M. J. Kellner, A. L. Tan, L. M. Paul, L. A. Parham, K. F. Garcia, K. G. Barnes, B. Chak, A. Mondini, M. L. Nogueira, S. Isern, S. F. Michael, I. Lorenzana, N. L. Yozwiak, B. L. MacInnis, I. Bosch, L. Gehrke, F. Zhang, P. C. Sabeti, Science 2018, 360, 444–448.
- 38R. Bruch, J. Baaske, C. Chatelle, M. Meirich, S. Madlener, W. Weber, C. Dincer, G. A. Urban, Adv. Mater. 2019, 31, 1905311.
- 39G. Zhong, H. Wang, Y. Li, M. H. Tran, M. Farzan, Nat. Chem. Biol. 2017, 13, 839–841.
- 40H. R. Kempton, L. E. Goudy, K. S. Love, L. S. Qi, Mol. Cell 2020, 78, 184–191.
- 41S. O. Kelley, C. A. Mirkin, D. R. Walt, R. F. Ismagilov, M. Toner, E. H. Sargent, Nat. Nanotechnol. 2014, 9, 969–980.
- 42W. Yang, H. Yu, O. Alkhamis, Y. Liu, J. Canoura, F. Fu, Y. Xiao, Nucleic Acids Res. 2019, 47, e71.
- 43Y. Xiong, J. Zhang, Z. Yang, Q. Mou, Y. Ma, Y. Xiong, Y. Lu, J. Am. Chem. Soc. 2020, 142, 207–213.
- 44D. Kern, E. R. Zuiderweg, Curr. Opin. Struct. Biol. 2003, 13, 748–757.
- 45N. M. Goodey, S. J. Benkovic, Nat. Chem. Biol. 2008, 4, 474–482.
- 46M. Liang, Z. Li, W. Wang, J. Liu, L. Liu, G. Zhu, L. Karthik, M. Wang, K.-F. Wang, Z. Wang, J. Yu, Y. Shuai, J. Yu, L. Zhang, Z. Yang, C. Li, Q. Zhang, T. Shi, L. Zhou, F. Xie, H. Dai, X. Liu, J. Zhang, G. Liu, Y. Zhuo, B. Zhang, C. Liu, S. Li, X. Xia, Y. Tong, Y. Liu, G. Alterovitz, G.-Y. Tan, L.-X. Zhang, Nat. Commun. 2019, 10, 3672.
- 47N. D. Taylor, A. S. Garruss, R. Moretti, S. Chan, M. A. Arbing, D. Cascio, J. K. Rogers, F. J. Isaacs, S. Kosuri, D. Baker, S. Fields, G. M. Church, S. Raman, Nat. Methods 2016, 13, 177–183.
- 48
- 48aY. Li, L. Liu, G. Liu, Trends Biotechnol. 2019, 37, 792–795;
- 48bY. Li, S. Li, J. Wang, G. Liu, Trends Biotechnol. 2019, 37, 730–743.
- 49R. Hajian, S. Balderston, T. Tran, T. deBoer, J. Etienne, M. Sandhu, N. A. Wauford, J.-Y. Chung, J. Nokes, M. Athaiya, J. Paredes, R. Peytavi, B. Goldsmith, N. Murthy, I. M. Conboy, K. Aran, Nat. Biomed. Eng. 2019, 3, 427–437.
- 50W. Xu, T. Jin, Y. Dai, C. C. Liu, Biosens. Bioelectron. 2020, 155, 112100.
- 51M. A. English, L. R. Soenksen, R. V. Gayet, H. de Puig, N. M. Angenent-Mari, A. S. Mao, P. Q. Nguyen, J. J. Collins, Science 2019, 365, 780–785.
- 52M. H. Hanewich-Hollatz, Z. Chen, L. M. Hochrein, J. Huang, N. A. Pierce, ACS Cent. Sci. 2019, 5, 1241–1249.
- 53M. R. O′Connell, B. L. Oakes, S. H. Sternberg, A. East-Seletsky, M. Kaplan, J. A. Doudna, Nature 2014, 516, 263–266.
- 54K. Pardee, A. A. Green, T. Ferrante, D. E. Cameron, A. DaleyKeyser, P. Yin, J. J. Collins, Cell 2014, 159, 940–954.