Identification of Chemokine Ligands by Biochemical Fragmentation and Simulated Peptide Evolution
Graphical Abstract
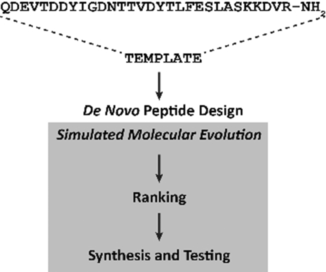
Fragmentation of the chemokine-receptor CCL19/CCR7 binding domain led to a hexapeptide as a template for de novo peptide design. Computer-generated peptides were characterized in terms of their binding to CCl19. Potent ligands were identified as starting points for developing innovative protein–protein interaction modulators.
Abstract
Short linear peptides can overcome certain limitations of small molecules for targeting protein–protein interactions (PPIs). Herein, the interaction between the human chemokine CCL19 with chemokine receptor CCR7 was investigated to obtain receptor-derived CCL19-binding peptides. After identifying a linear binding site of CCR7, five hexapeptides binding to CCL19 in the low micromolar to nanomolar range were designed, guided by pharmacophore and lipophilicity screening of computationally generated peptide libraries. The results corroborate the applicability of the computational approach and the chosen selection criteria to obtain short linear peptides mimicking a protein–protein interaction site.