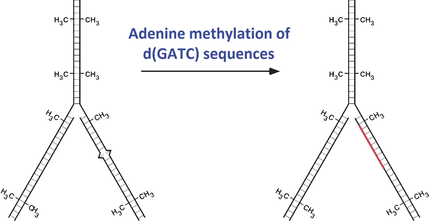
Mechanisms in E. coli and Human Mismatch Repair (Nobel Lecture)†
Corresponding Author
Prof. Paul Modrich
Howard Hughes Medical Institute and Department of Biochemistry, Duke University, Medical Center, Durham, NC, 27710 USA
Search for more papers by this authorCorresponding Author
Prof. Paul Modrich
Howard Hughes Medical Institute and Department of Biochemistry, Duke University, Medical Center, Durham, NC, 27710 USA
Search for more papers by this authorCopyright© The Nobel Foundation 2015. We thank the Nobel Foundation, Stockholm, for permission to print this lecture.
Graphical Abstract
DNA molecules are not completely stable, they are subject to chemical or photochemical damage and errors that occur during DNA replication resulting in mismatched base pairs. Through mechanistic studies Paul Modrich showed how replication errors are corrected by strand-directed mismatch repair in Escherichia coli and human cells.
References
- 1“A mechanism for gene conversion in fungi”: R. Holliday, Genet. Res. 1964, 5, 282–304.
- 2“Chromosome breakage accompanying genetic recombination in bacteriophage”: M. Meselson, J. J. Weigle, Proc. Natl. Acad. Sci. USA 1961, 47, 857–868.
- 3“Genetic studies of recombining DNA in pneumococcal transformation”: H. Ephrussi-Taylor, T. C. Gray, J. Gen. Physiol. 1966, 49, 211–231.
- 4“Genetic consequences of transfection with heteroduplex bacteriophage lambda DNA”: R. L. White, M. S. Fox, Mol. Gen. Genet. 1975, 141, 163–171.
- 5“Mismatch repair in heteroduplex DNA”: J. Wildenberg, M. Meselson, Proc. Natl. Acad. Sci. USA 1975, 72, 2202–2206.
- 6“Repair tracts in mismatched DNA heteroduplexes”: R. Wagner, M. Meselson, Proc. Natl. Acad. Sci. USA 1976, 73, 4135–4139.
- 7“Postreplicative mismatch repair”: J. Jiricny, Cold Spring Harbor Perspect. Biol. 2013, 5, a 012633.
- 8“Effects of high levels of DNA adenine methylation on methyl-directed mismatch repair in Escherichia coli”: P. J. Pukkila, J. Peterson, G. Herman, P. Modrich, M. Meselson, Genetics 1983, 104, 571–582.
- 9“Escherichia coli mutants uvrD uvrE deficient in gene conversion of lambda heteroduplexes”: P. Nevers, H. Spatz, Mol. Gen. Genet. 1975, 139, 233–243.
- 10“Bromouracil mutagenesis and mismatch repair in mutator strains of Escherichia coli”: B. Rydberg, Mutat. Res. 1978, 52, 11–24.
- 11“Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction”: B. W. Glickman, M. Radman, Proc. Natl. Acad. Sci. USA 1980, 77, 1063–1067.
- 12“Restriction endonuclease B and f1 heteroduplex DNA”: G. F. Vovis, K. Horiuchi, N. Hartman, N. D. Zinder, Nat. New Biol. 1973, 246, 13–16.
- 13“Genetic studies with heteroduplex DNA of bacteriophage f1. Asymmetric segregation, base correction, and implications for the mechanism of genetic recombination”: V. Enea, G. F. Vovis, N. D. Zinder, J. Mol. Biol. 1975, 96, 495–509.
- 14“Methyl-directed repair of DNA base-pair mismatches in vitro”: A. L. Lu, S. Clark, P. Modrich, Proc. Natl. Acad. Sci. USA 1983, 80, 4639–4643.
- 15“Repair of DNA base-pair mismatches in extracts of Escherichia coli”: A. L. Lu, K. Welsh, S. Clark, S. S. Su, P. Modrich, Cold Spring Harbor Symp. Quant. Biol. 1984, 49, 589–596.
- 16“Mispair specificity of methyl-directed DNA mismatch correction in vitro”: S.-S. Su, R. S. Lahue, K. G. Au, P. Modrich, J. Biol. Chem. 1988, 263, 6829–6835.
- 17“Requirement for d(GATC) sequences in Escherichia coli mutHLS mismatch correction”: R. S. Lahue, S. S. Su, P. Modrich, Proc. Natl. Acad. Sci. USA 1987, 84, 1482–1486.
- 18“Gap formation is associated with methyl-directed mismatch correction under conditions of restricted DNA synthesis”: S. S. Su, M. Grilley, R. Thresher, J. Griffith, P. Modrich, Genome 1989, 31, 104–111.
- 19“Bidirectional excision in methyl-directed mismatch repair”: M. Grilley, J. Griffith, P. Modrich, J. Biol. Chem. 1993, 268, 11830–11837.
- 20“Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs”: S.-S. Su, P. Modrich, Proc. Natl. Acad. Sci. USA 1986, 83, 5057–5061.
- 21“Isolation and characterization of the Escherichia coli mutH gene product”: K. M. Welsh, A. L. Lu, S. Clark, P. Modrich, J. Biol. Chem. 1987, 262, 15624–15629.
- 22“Isolation and characterization of the Escherichia coli mutL gene product”: M. Grilley, K. M. Welsh, S.-S. Su, P. Modrich, J. Biol. Chem. 1989, 264, 1000–1004.
- 23“DNA mismatch correction in a defined system”: R. S. Lahue, K. G. Au, P. Modrich, Science 1989, 245, 160–164.
- 24“The E. coli uvrD gene product is DNA helicase II”: I. D. Hickson, H. M. Arthur, D. Bramhill, P. T. Emmerson, Mol. Gen. Genet. 1983, 190, 265–270.
- 25“The crystal structure of DNA mismatch repair protein MutS binding to a G-T mismatch”: M. H. Lamers, A. Perrakis, J. H. Enzlin, H. H. Winterwerp, N. de Wind, T. K. Sixma, Nature 2000, 407, 711–717.
- 26“Methyl-directed mismatch repair is bidirectional”: D. L. Cooper, R. S. Lahue, P. Modrich, J. Biol. Chem. 1993, 268, 11823–11829.
- 27“In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair”: V. Burdett, C. Baitinger, M. Viswanathan, S. T. Lovett, P. Modrich, Proc. Natl. Acad. Sci. USA 2001, 98, 6765–6770.
- 28“Redundant exonuclease involvement in Escherichia coli methyl-directed mismatch repair”: M. Viswanathan, V. Burdett, C. Baitinger, P. Modrich, S. T. Lovett, J. Biol. Chem. 2001, 276, 31053–31058.
- 29“Exonuclease VII of Escherichia coli: mechanism of action”: J. W. Chase, C. C. Richardson, J. Biol. Chem. 1974, 249, 4553–4561.
- 30“Initiation of methyl-directed mismatch repair”: K. G. Au, K. Welsh, P. Modrich, J. Biol. Chem. 1992, 267, 12142–12148.
- 31“Mismatch, MutS, MutL, and helicase II,-dependent unwinding from the single-strand break of an incised heteroduplex”: V. Dao, P. Modrich, J. Biol. Chem. 1998, 273, 9202–9207.
- 32“MutS and MutL activate DNA helicase II, in a mismatch-dependent manner”: M. Yamaguchi, V. Dao, P. Modrich, J. Biol. Chem. 1998, 273, 9197–9201.
- 33“Involvement of the beta clamp in methyl-directed mismatch repair in vitro”: A. Pluciennik, V. Burdett, O. Lukianova, M. O'Donnell, P. Modrich, J. Biol. Chem. 2009, 284, 32782–32791.
- 34“DNA Mismatch Repair”: T. A. Kunkel, D. A. Erie, Annu. Rev. Biochem. 2005, 74, 681–710.
- 35“DNA mismatch repair: functions and mechanisms”: R. R. Iyer, A. Pluciennik, V. Burdett, P. L. Modrich, Chem. Rev. 2006, 106, 302–323.
- 36“Mismatch Repair”: R. Fishel, J. Biol. Chem. 2015, 290, 26395–26403.
- 37“Mismatch correction catalyzed by cell-free extracts of Saccharomyces cerevisiae”: C. Muster-Nassal, R. Kolodner, Proc. Natl. Acad. Sci. USA 1986, 83, 7618–7622.
- 38“DNA mismatch repair detected in human cell extracts”: P. M. Glazer, S. N. Sarkar, G. E. Chisholm, W. C. Summers, Mol. Cell. Biol. 1987, 7, 218–224.
- 39“Different base/base mispairs are corrected with different efficiencies and specificities in monkey kidney cells”: T. C. Brown, J. Jiricny, Cell 1988, 54, 705–711.
- 40“Mismatch repair involving localized DNA synthesis in extracts of Xenopus eggs”: P. Brooks, C. Dohet, G. Almouzni, M. Mechali, M. Radman, Proc. Natl. Acad. Sci. USA 1989, 86, 4425–4429.
- 41“Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines”: J. Holmes, S. Clark, P. Modrich, Proc. Natl. Acad. Sci. USA 1990, 87, 5837–5841.
- 42“Clues to the pathogenesis of familial colorectal cancer”: L. A. Aaltonen, P. Peltomäki, F. S. Leach, P. Sistonen, L. Pylkkänen, J.-P. Mecklin, H. Järvinen, S. M. Powell, J. Jen, S. R. Hamilton, G. M. Petersen, K. W. Kinzler, B. Vogelstein, A. de la Chapelle, Science 1993, 260, 812–816.
- 43“Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis”: Y. Ionov, M. A. Peinado, S. Malkhosyan, D. Shibata, M. Perucho, Nature 1993, 363, 558–561.
- 44“Role of DNA mismatch repair defects in the pathogenesis of human cancer”: P. Peltomaki, J. Clin. Oncol. 2003, 21, 1174–1179.
- 45“Inherited susceptibility to colorectal cancer”: P. T. Rowley, Annu. Rev. Med. 2005, 56, 539–554.
- 46“High frequencies of short frameshifts in poly-CA/TG tandem repeats borne by bacteriophage M13 in Escherichia coli K-12”: G. Levinson, G. A. Gutman, Nucleic Acids Res. 1987, 15, 5323–5338.
- 47“Hypermutability and mismatch repair deficiency in RER+ tumor cells”: R. Parsons, G. M. Li, M. J. Longley, W. H. Fang, N. Papadopoulos, J. Jen, A. de la Chapelle, K. W. Kinzler, B. Vogelstein, P. Modrich, Cell 1993, 75, 1227–1236.
- 48“Isolation of an hMSH2⋅p160 heterodimer that restores mismatch repair to tumor cells”: J. T. Drummond, G.-M. Li, M. J. Longley, P. Modrich, Science 1995, 268, 1909–1912.
- 49“Restoration of mismatch repair to nuclear extracts of H6 colorectal tumor cells by a heterodimer of human MutL homologs”: G. M. Li, P. Modrich, Proc. Natl. Acad. Sci. USA 1995, 92, 1950–1954.
- 50“Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers”: M. L. Veigl, L. Kasturi, J. Olechnowicz, A. H. Ma, J. D. Lutterbaugh, S. Periyasamy, G. M. Li, J. Drummond, P. L. Modrich, W. D. Sedwick, S. D. Markowitz, Proc. Natl. Acad. Sci. USA 1998, 95, 8698–8702.
- 51“MSH6, a Saccharomyces cerevisiae protein that binds to mismatches as a heterodimer with MSH2”: I. Iaccarino, F. Palombo, J. Drummond, N. F. Totty, J. J. Hsuan, P. Modrich, J. Jiricny, Curr. Biol. 1996, 6, 484–486.
- 52“Isolation of hMutSbeta from human cells and comparison of the mismatch repair specificities of hMutSbeta and hMutSalpha”: J. Genschel, S. J. Littman, J. T. Drummond, P. Modrich, J. Biol. Chem. 1998, 273, 19895–19901.
- 53“Identification of mismatch repair genes and their role in the development of cancer”: R. Fishel, R. D. Kolodner, Curr. Opin. Genet. Dev. 1995, 5, 382–395.
- 54“Lessons from hereditary colorectal cancer”: K. W. Kinzler, B. Vogelstein, Cell 1996, 87, 159–170.
- 55“Increased mutation rate at the hprt locus accompanies microsatellite instability in colon cancer”: J. R. Eshleman, E. Z. Lang, G. K. Bowerfind, R. Parsons, B. Vogelstein, J. K. Willson, M. L. Veigl, W. D. Sedwick, S. D. Markowitz, Oncogene 1995, 10, 33–37.
- 56“Mutation rate at the hprt locus in human cancer cell lines with specific mismatch repair-gene defects”: W. E. Glaab, K. R. Tindall, Carcinogenesis 1997, 18, 1–8.
- 57“Functional overlap in mismatch repair by human MSH3 and MSH6”: A. Umar, J. I. Risinger, W. E. Glaab, K. R. Tindall, J. C. Barrett, T. A. Kunkel, Genetics 1998, 148, 1637–1646.
- 58“Mutator pathways unleashed by epigenetic silencing in human cancer”: F. V. Jacinto, M. Esteller, Mutagenesis 2007, 22, 247–253.
- 59“hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6”: S. Acharya, T. Wilson, S. Gradia, M. F. Kane, S. Guerrette, G. T. Marsischky, R. Kolodner, R. Fishel, Proc. Natl. Acad. Sci. USA 1996, 93, 13629–13634.
- 60“Binding of insertion/deletion DNA mismatches by the heterodimer of yeast mismatch repair proteins MSH2 and MSH3”: Y. Habraken, P. Sung, L. Prakash, S. Prakash, Curr. Biol. 1996, 6, 1185–1187.
- 61“hMutSb, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA”: F. Palombo, I. Iaccarino, E. Nakajima, M. Ikejima, T. Shimada, J. Jiricny, Curr. Biol. 1996, 6, 1181–1184.
- 62“A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction”: P. Szankasi, G. R. Smith, Science 1995, 267, 1166–1169.
- 63“Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2”: D. X. Tishkoff, A. L. Boerger, P. Bertrand, N. Filosi, G. M. Gaida, M. F. Kane, R. D. Kolodner, Proc. Natl. Acad. Sci. USA 1997, 94, 7487–7492.
- 64“Human exonuclease I is required for 5′ and 3′ mismatch repair”: J. Genschel, L. R. Bazemore, P. Modrich, J. Biol. Chem. 2002, 277, 13302–13311.
- 65“The evolutionarily conserved zinc finger motif in the largest subunit of human replication protein A is required for DNA replication and mismatch repair but not for nucleotide excision repair”: Y. L. Lin, M. K. Shivji, C. Chen, R. Kolodner, R. D. Wood, A. Dutta, J. Biol. Chem. 1998, 273, 1453–1461.
- 66“Partial reconstitution of human DNA mismatch repair in vitro: characterization of the role of human replication protein A”: C. Ramilo, L. Gu, S. Guo, X. Zhang, S. M. Patrick, J. J. Turchi, G. M. Li, Mol. Cell. Biol. 2002, 22, 2037–2046.
- 67“Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis”: A. Umar, A. B. Buermeyer, J. A. Simon, D. C. Thomas, A. B. Clark, R. M. Liskay, T. A. Kunkel, Cell 1996, 87, 65–73.
- 68“A defined human system that supports bidirectional mismatch-provoked excision”: L. Dzantiev, N. Constantin, J. Genschel, R. R. Iyer, P. M. Burgers, P. Modrich, Mol. Cell 2004, 15, 31–41.
- 69“DNA polymerase delta is required for human mismatch repair in vitro”: M. J. Longley, A. J. Pierce, P. Modrich, J. Biol. Chem. 1997, 272, 10917–10921.
- 70“Human mismatch repair: Reconstitution of a nick-directed bidirectional reaction”: N. Constantin, L. Dzantiev, F. A. Kadyrov, P. Modrich, J. Biol. Chem. 2005, 280, 39752–39761.
- 71“Mechanism of 5′-directed excision in human mismatch repair”: J. Genschel, P. Modrich, Mol. Cell 2003, 12, 1077–1086.
- 72“Reconstitution of 5′-directed human mismatch repair in a purified system”: Y. Zhang, F. Yuan, S. R. Presnell, K. Tian, Y. Gao, A. E. Tomkinson, L. Gu, G. M. Li, Cell 2005, 122, 693–705.
- 73“Functions of MutL{alpha}, RPA, and HMGB1 in 5′-directed mismatch repair”: J. Genschel, P. Modrich, J. Biol. Chem. 2009, 284, 21536–21544.
- 74“PARP-1 enhances the mismatch-dependence of 5′-directed excision in human mismatch repair in vitro”: Y. Liu, F. A. Kadyrov, P. Modrich, DNA Repair 2011, 10, 1145–1153.
- 75“Endonucleolytic function of MutLalpha in human mismatch repair”: F. A. Kadyrov, L. Dzantiev, N. Constantin, P. Modrich, Cell 2006, 126, 297–308.
- 76“Saccharomyces cerevisiae MutLα is a mismatch repair endonuclease”: F. A. Kadyrov, S. F. Holmes, M. E. Arana, O. A. Lukianova, M. O'Donnell, T. A. Kunkel, P. Modrich, J. Biol. Chem. 2007, 282, 37181–37190.
- 77“PMS2 endonuclease activity has distinct biological functions and is essential for genome maintenance”: J. M. van Oers, S. Roa, U. Werling, Y. Liu, J. Genschel, H. Hou, Jr., R. S. Sellers, P. Modrich, M. D. Scharff, W. Edelmann, Proc. Natl. Acad. Sci. USA 2010, 107, 13384–13389.
- 78“Mismatch repair in Gram-positive bacteria”: J. S. Lenhart, M. C. Pillon, A. Guarne, J. S. Biteen, L. A. Simmons, Res. Microbiol. 2016, in press.
- 79“PCNA function in the activation and strand direction of MutLalpha endonuclease in mismatch repair”: A. Pluciennik, L. Dzantiev, R. R. Iyer, N. Constantin, F. A. Kadyrov, P. Modrich, Proc. Natl. Acad. Sci. USA 2010, 107, 16066–16071.
- 80“hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci”: H. E. Kleczkowska, G. Marra, T. Lettieri, J. Jiricny, Genes Dev. 2001, 15, 724–736.
- 81“Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates”: H. Hombauer, C. S. Campbell, C. E. Smith, A. Desai, R. D. Kolodner, Cell 2011, 147, 1040–1053.
- 82“Replication clamps and clamp loaders”: M. Hedglin, R. Kumar, S. J. Benkovic, Cold Spring Harbor Perspect. Biol. 2013, 5, a 010165.
- 83“Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria”: J.-P. Claverys, S. A. Lacks, Microbiol. Rev. 1986, 50, 133–165.
- 84“Exonuclease 1 preferentially repairs mismatches generated by DNA polymerase alpha”: S. E. Liberti, A. A. Larrea, T. A. Kunkel, DNA Repair 2013, 12, 92–96.