A Phosphorylation-Induced Turn Defines the Alzheimer’s Disease AT8 Antibody Epitope on the Tau Protein†
We thank Drs. J. Götz, B. Chambraud, and E. E. Baulieu for helpful discussion, and Dr. L. Peyriga for help with setting up the NMR experiments on the 800 MHz machine (LISBP, Toulouse, France). The 900 MHz NMR facility was supported by the CNRS (TGIR RMN THC, FR-3050, France), University of Lille 1, the European community (EDRF), and the Région Nord-Pas de Calais. Part of this work was financed by the ANR (program MALZ-TAF) and LabEx Distalz grant (France). The computational work was funded by a Curtin Early Research fellowship. This work was supported by resources provided by the Pawsey Supercomputing Centre with funding from the Australian Government and the Government of Western Australia.
Graphical Abstract
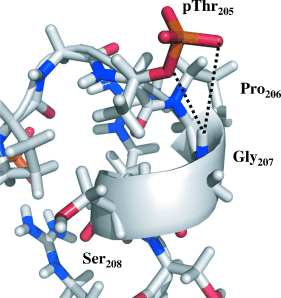
The AT8 epitope of the phosphorylated tau protein was mapped by NMR spectroscopy, and its defining structural features were derived by a combination of NMR analyses and molecular dynamics. A particular turn conformation is stabilized by a hydrogen bond of the phosphorylated Thr205 residue (pThr205) to the amide proton of Gly207.
Abstract
Post mortem biochemical staging of Alzheimer’s disease is currently based on immunochemical analysis of brain slices with the AT8 antibody. The epitope of AT8 is described around the pSer202/pThr205 region of the hyperphosphorylated form of the neuronal protein tau. In this study, NMR spectroscopy was used to precisely map the AT8 epitope on phosphorylated tau, and derive its defining structural features by a combination of NMR analyses and molecular dynamics. A particular turn conformation is stabilized by a hydrogen bond of the phosphorylated Thr205 residue to the amide proton of Gly207, and is further stabilized by the two Arg residues opposing the pSer202/pThr205.