Crystal Structure of a Spin-Labeled, Channel-Forming Alamethicin Analogue†
Marco Crisma Dr.
Istituto di Chimica Biomolecolare, CNR, Dipartimento di Scienze Chimiche, Università degli Studi di Padova via Marzolo 1, 35131 Padova, Italy, Fax: (+39) 049-827-5239
Search for more papers by this authorCristina Peggion Dr.
Istituto di Chimica Biomolecolare, CNR, Dipartimento di Scienze Chimiche, Università degli Studi di Padova via Marzolo 1, 35131 Padova, Italy, Fax: (+39) 049-827-5239
Search for more papers by this authorChiara Baldini
Istituto di Chimica Biomolecolare, CNR, Dipartimento di Scienze Chimiche, Università degli Studi di Padova via Marzolo 1, 35131 Padova, Italy, Fax: (+39) 049-827-5239
Search for more papers by this authorElizabeth J. MacLean Dr.
CCLRC Daresbury Laboratory, Daresbury, Warrington, Cheshire WA4 4AD, UK
Search for more papers by this authorNatascia Vedovato
Dipartimento di Biologia, Sezione di Fisiologia e Biofisica, Università degli Studi di Ferrara, 44100 Ferrara, Italy
Search for more papers by this authorGiorgio Rispoli Prof.
Dipartimento di Biologia, Sezione di Fisiologia e Biofisica, Università degli Studi di Ferrara, 44100 Ferrara, Italy
Search for more papers by this authorClaudio Toniolo Prof.
Istituto di Chimica Biomolecolare, CNR, Dipartimento di Scienze Chimiche, Università degli Studi di Padova via Marzolo 1, 35131 Padova, Italy, Fax: (+39) 049-827-5239
Search for more papers by this authorMarco Crisma Dr.
Istituto di Chimica Biomolecolare, CNR, Dipartimento di Scienze Chimiche, Università degli Studi di Padova via Marzolo 1, 35131 Padova, Italy, Fax: (+39) 049-827-5239
Search for more papers by this authorCristina Peggion Dr.
Istituto di Chimica Biomolecolare, CNR, Dipartimento di Scienze Chimiche, Università degli Studi di Padova via Marzolo 1, 35131 Padova, Italy, Fax: (+39) 049-827-5239
Search for more papers by this authorChiara Baldini
Istituto di Chimica Biomolecolare, CNR, Dipartimento di Scienze Chimiche, Università degli Studi di Padova via Marzolo 1, 35131 Padova, Italy, Fax: (+39) 049-827-5239
Search for more papers by this authorElizabeth J. MacLean Dr.
CCLRC Daresbury Laboratory, Daresbury, Warrington, Cheshire WA4 4AD, UK
Search for more papers by this authorNatascia Vedovato
Dipartimento di Biologia, Sezione di Fisiologia e Biofisica, Università degli Studi di Ferrara, 44100 Ferrara, Italy
Search for more papers by this authorGiorgio Rispoli Prof.
Dipartimento di Biologia, Sezione di Fisiologia e Biofisica, Università degli Studi di Ferrara, 44100 Ferrara, Italy
Search for more papers by this authorClaudio Toniolo Prof.
Istituto di Chimica Biomolecolare, CNR, Dipartimento di Scienze Chimiche, Università degli Studi di Padova via Marzolo 1, 35131 Padova, Italy, Fax: (+39) 049-827-5239
Search for more papers by this authorProvision of time on the Small Molecule Crystallography Service at the CCLRC Daresbury Laboratory through support by the European Community Research Infrastructure Action under the FP6 “Structuring the European Research Area” Programme (through the Integrated Infrastructure Initiative “Integrating Activity on Synchrotron and Free Electron Laser Science”) is acknowledged.
Graphical Abstract
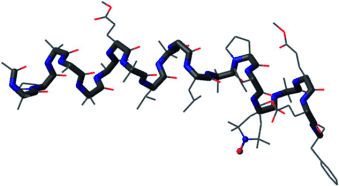
Crystal clear: A detailed conformational characterization of a synthetic analogue of the peptide antibiotic alamethicin (see structure; C gray, N blue, O red) has been achieved by X-ray diffraction. The high-resolution structure of this analogue, which incorporates a spin probe, paves the way for a better understanding of the mode of action of alamethicin.
Supporting Information
Supporting information for this article is available on the WWW under http://www.wiley-vch.de/contents/jc_2002/2007/z604417_s.pdf or from the author.
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
References
- 1
- 1aE. Benedetti, A. Bavoso, B. Di Blasio, V. Pavone, C. Pedone, C. Toniolo, G. M. Bonora, Proc. Natl. Acad. Sci. USA 1982, 79, 7951–7954;
- 1bH. Brückner, H. Graf, Experientia 1983, 39, 528–530;
- 1cL. Whitmore, J. K. Chugh, C. F. Snook, B. A. Wallace, J. Pept. Sci. 2003, 9, 663–665.
- 2J. Kirschbaum, C. Krause, R. K. Winzheimer, H. Brückner, J. Pept. Sci. 2003, 9, 799–809.
- 3
- 3aR. Nagaraj, P. Balaram, Acc. Chem. Res. 1981, 14, 356–362;
- 3bG. Boheim, W. Hanke, G. Jung, Biophys. Struct. Mech. 1983, 9, 181–191;
- 3cT. Rink, H. Bartel, G. Jung, Eur. Biophys. J. 1994, 23, 155–165;
- 3dM. S. P. Sansom, Prog. Biophys. Mol. Biol. 1991, 55, 139–235;
- 3eB. Bechinger, J. Membr. Biol. 1997, 156, 197–211;
- 3fJ. K. Chugh, B. A. Wallace, Biochem. Soc. Trans. 2001, 29, 565–570.
- 4C. Peggion, M. Jost, C. Baldini, F. Formaggio, C. Toniolo, Chem. Biodiversity, in press.
- 5M. Crisma, J. R. Deschamps, C. George, J. L. Flippen-Anderson, B. Kaptein, Q. B. Broxterman, A. Moretto, S. Oancea, M. Jost, F. Formaggio, C. Toniolo, J. Pept. Res. 2005, 65, 564–579.
- 6M. Barranger Mathys, D. S. Cafiso, Biochemistry 1996, 35, 498–505.
- 7R. O. Fox, F. M. Richards, Nature 1982, 300, 325–330.
- 8
- 8aJ. K. Chugh, H. Brückner, B. A. Wallace, Biochemistry 2002, 41, 12934–12941;
- 8bI. L. Karle, J. L. Flippen-Anderson, S. Agarwalla, P. Balaram, Proc. Natl. Acad. Sci. USA 1991, 88, 5307–5311;
- 8cI. L. Karle, M. A. Perozzo, V. K. Mishra, P. Balaram, Proc. Natl. Acad. Sci. USA 1998, 95, 5501–5504;
- 8dC. F. Snook, G. A. Woolley, G. Oliva, V. Pattabhi, S. F. Wood, T. L. Blundell, B. A. Wallace, Structure 1998, 6, 783–792;
- 8eG. Bunkoczi, M. Schiell, L. Vertesi, G. M. Sheldrick, J. Pept. Sci. 2003, 9, 745–752;
- 8fM. Kronen, H. Görls, H.-H. Nguyen, S. Reissmann, M. Bohl, J. Sühnel, U. Gräfe, J. Pept. Sci. 2003, 9, 729–744;
- 8gC. Toniolo, C. Peggion, M. Crisma, F. Formaggio, X. Shui, D. S. Eggleston, Nat. Struct. Biol. 1994, 1, 908–914;
- 8hM. Crisma, A. Barazza, F. Formaggio, B. Kaptein, Q. B. Broxterman, J. Kamphuis, C. Toniolo, Tetrahedron 2001, 57, 2813–2825.
- 9Crystallographic data: 2 C101H164N21O28⋅3 H2O, Mr=4295.1, 0.40×0.06×0.02 mm3, monoclinic, space group C2, a=39.767(4), b=20.192(3), c=32.655(3) Å, β=116.091(2)°, V=23 549(5) Å3, Z=4, ρcalcd=1.211 Mg m−3, μ=0.090 mm−1, λ=0.6868 Å, T=150 K, 2 θmax=42.4°. In total, 68 928 reflections were collected, of which 28 257 were independent (Rint=0.041) and employed for refinement, except for a 5 % fraction, which was reserved for Rfree calculation. Data/restraints/parameters: 26 845/851/2725, R1=0.117 (F≥4 σ(F)), wR2=0.305 (all data), min/max residual electron density: −0.38/0.95 e Å−3. Data were collected at the microcrystal diffraction facility on Station 9.8 of the Synchrotron Radiation Source, CCLRC Daresbury Laboratory. The structure was solved ab initio using SHELXD,[12] and refined by least-squares procedures using SHELXL-97.[13] Details of the structure solution and refinement, along with tables of torsion angles and hydrogen-bond parameters, may be found in the Supporting Information. CCDC-604861 contains the supplementary crystallographic data for this paper. These data can be obtained free of charge from The Cambridge Crystallographic Data Centre via www.ccdc.cam.ac.uk/data_request/cif.
- 10C. Toniolo, Crit. Rev. Biochem. 1980, 9, 1–44.
- 11D. P. Tieleman, M. S. P. Sansom, H. J. C. Berendsen, Biophys. J. 1999, 76, 40–49.
- 12G. M. Sheldrick, H. A. Hauptmann, C. M. Weeks, R. Miller, I. Usón in International Tables for Crystallography, Vol. F (Eds.: ), Kluwer Academic, Dordrecht, 2001, pp. 333–351.
- 13G. M. Sheldrick, SHELXL-97, Program for the Refinement of Crystal Structures, University of Göttingen, Göttingen (Germany), 1997.