Bacterial communities in the nasal passage of postviral olfactory dysfunction patients
Abstract
Key points
-
Bacterial composition is uniform in the sinuses of postviral olfactory dysfunction patients.
-
Significant reduction of genus Corynebacterium in PVOD patients compared to controls.
1 INTRODUCTION
Persistent olfactory dysfunction may occur with viral or bacterial infections of the upper respiratory tract.1, 2 Other factors such as anatomical structure, genetics, neurodegenerative disorders, and age could also impact olfaction.2
Our own study3 and that of Koskinen at al.4 have shown some evidence of differences in the nasal microbiota composition between anosmia, hyposmia, and normosmia cohorts. However, both these studies used different sampling sites and it remains unclear whether the microbiota at different sites in the nose of patients with olfactory dysfunction varies. The histology of these two sites differ structurally and functionally, and one would expect the microbes colonizing these sites to be different.5
Although our previous study focused on only one sampling site (middle meatus), this study primarily investigated differences in the microbiome in the anterior nares, middle meatus, and olfactory cleft of postviral olfactory dysfunction (PVOD) patients and healthy controls.
2 PATIENTS AND METHODS
2.1 Patient recruitment
The study was authorized by the ethics committee at the medical faculty at the Technical University of Dresden (ethics #EK 233072018). Written consent was obtained from all participants.
Twenty PVOD and 20 healthy control participants were recruited from the Carl Gustav Carus University Hospital between March and October 2021. A detailed history regarding the genesis of the olfactory loss and a thorough otorhinolaryngological examination was performed in the consultation.
2.2 Sample collection
Pairs of sterile rayon-tipped swabs (Copan, #170KS01) were collected by an ear, nose, and throat (ENT) specialist using an endoscope for visualization, on either the left or right side at three sites—on the olfactory cleft, the middle meatus, and the inferior turbinate.
The swabs were stored at −20°C in 1 mL RNAlater for sample preservation. Once all samples were collected, they were shipped on dry ice to University of Colorado Anschutz Medical Center, Denver, CO, USA. Samples were immediately placed in a −80°C freezer.
2.3 Threshold, discrimination, and identification smell test
The threshold, discrimination, and identification (TDI) standardized smell test is named after the initials of the three tests: olfactory threshold (T), differentiation (D), and identification ability (I) of scents. All participants were tested as described using “Sniffin' Sticks.”6
A final score across all three tests (TDI) of ≤16.5 are considered anosmic, >30.5 are considered normosmic, and hyposmia as a score between these two values.
2.4 DNA extraction, polymerase chain reaction, and sequencing
Total DNA was extracted from swab samples using the QIAamp PowerFecal DNA kit (Qiagen Inc, Carlsbad, CA, USA) following the manufacturer's protocol. The V1V2 region of the bacterial 16S ribosomal RNA (rRNA) genes were amplified, normalized, quantified, and sequenced as described.7
A total of six negative control samples (4 polymerase chain reaction [PCR] negative, 2 DNA extraction negative) were included for sequencing. Raw sequence reads are stored on a National Center for Biotechnology Information (NCBI) database (bio-project PRJNA921889).
2.5 Bioinformatics and statistical analysis
Sequence data were processed according to the DADA2 pipeline version 1.168 in R statistical program version 4.1.29 as described3 Dunn's test or Kruskal-Wallis test with Benjamini-Hochberg p value correction were used to test differences between groups.
3 RESULTS
No significant differences were observed between PVOD and healthy control groups for all measured clinical variables. The participant information is presented in Table 1.
| Absolute and relative frequency | |||
|---|---|---|---|
| Characteristic | PVOD patients | Healthy controls | Overall |
| Gender, n (%) | |||
| Male | 7 | 7 | 14 (35%) |
| Female | 13 | 13 | 26 (65%) |
| Age (years) | |||
| Minimum–maximum | 20–76 | 22–84 | 20–84 |
| Mean | 45.6 ± 16.8 | 49.5 ± 19.9 | 47.5 ± 18.3 |
| Months between infection and the onset of smell loss | |||
| Minimum–maximum | 0–3 | ||
| Mean | 0.2 ± 0.7 | ||
| Immediate loss of smell after 0 months, n (%) | 17 (85) | ||
| Duration of odor loss in months | |||
| Minimum–maximum | 4–115 | ||
| Mean | 12.3 ± 24.5 | ||
| Postviral cause, n (%) | |||
| COVID-19 | 16 (80) | ||
| Other Infection | 4 (20) | ||
| Preexisting illness, n (%) | |||
| Overall | 10 (50) | 9 (45) | 19 (47.5) |
| Arterial hypertension | 1 (5) | 9 (45) | 10 (25) |
| Hypothyroidism | 3 (15) | 2 (10) | 5 (12.5) |
| Asthma | 4 (20) | 1 (5) | 5 (12.5) |
| Type 2 diabetes | 0 (0) | 4 (20) | 4 (10) |
| Allergies (including food, drugs, metal, latex, insects), n (%) | 13 (65) | 12 (60) | 25 (62.5) |
| Head operations | 11 (55) | 5 (25) | 18 (45) |
| Alcohol | 13 (65) | 17 (85) | 30 (75) |
| Influenza vaccination | 10 (50) | 10 (50) | 20 (50) |
| Corticosteroid nasal spray | 2 (5) | 0 | 2 (5) |
- Note: Patients with current or previous chronic nasal disease, <18 years of age, pregnancy, currently smoking, Parkinson's disease, Alzheimer's disease, or renal insufficiency were excluded. Subjects who had taken systemic antibiotics or glucocorticoids in the 4 weeks prior to recruitment were also excluded. Mean values are reported with standard deviation (±SD). The scale of the tests are from minimum 0 to maximum 10.
- Abbreviations: COVID-19, coronavirus disease 2019; PVOD, postviral olfactory dysfunction.
Fifteen subjects of the PVOD group were diagnosed with hyposmia and five with anosmia. The PVOD group achieved an average total TDI score of 20.28 ± 4.61 compared to the control group score of 35.45 ± 3.03.
3.1 Bacterial diversity and composition
No significant differences in bacterial diversity were observed between sampling locations or participants age or olfactory function. However, there was a trend for olfactory impaired individuals to have lower observed richness (controls = 61.9 ± 45.6 compared to PVOD = 42.6 ± 30.4, p = 0.06).
The dominating taxa at genus-level were Corynebacterium (relative abundance 36.7% ± 30.9%), Cutibacterium (21.9% ± 19.2%), and Staphylococcus (18.8% ± 21.4%) (Figure 1). This pattern was observed across all three sampling sites.
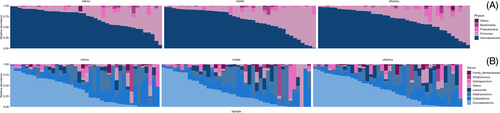
The PERMANOVA results showed that the majority (R2 = 84%, p = 0.001) of the variation observed in this study was due to interpersonal variability. Olfactory function (PVOD vs. healthy) accounted for a small fraction of the variation (R2 = 4.2%, p = 0.001). Age accounted for 12% (p = 0.001) of the variation. Other tested variables such as gender, steroid usage, and sample site did not contribute significantly to the variation in the bacterial community structure.
Corynebacterium The only amplicon seqeunce variant (ASV) with significant difference abundance between PVOD (relative abundance 2.7% ± 7.7%) and healthy controls (14.8% ± 18.9%) was Corynebacterium ASV3.
4 DISCUSSION
There are many biases that occur when conducting microbiome related studies, making it difficult to compare results across studies.10 Accordingly, this study set out to investigate the bacterial community composition at commonly sampled sinonasal sites in participants with olfactory impairment and healthy controls. The results of the present study show that the bacterial community diversity and composition do not differ between sample site locations in the sinuses.
The overwhelming interpersonal variation observed in this study makes it difficult to decipher any meaningful biological data from the results. However, the bacterial communities identified in this study are consistent with previous findings.3, 4 Our previous study3 and the study presented here both show a decrease in bacterial diversity in olfactory impaired individuals. There is also consistency between the two studies for a significant decrease in Corynebacterium species in PVOD individuals compared with healthy controls. We suggest future studies utilize quantitative measurements, such as droplet digital PCR (ddPCR), to measure the absolute abundances of this taxa.
ACKNOWLEDGMENTS
Open access publishing facilitated by The University of Auckland, as part of the Wiley - The University of Auckland agreement via the Council of Australian University Librarians.




