Molecular and phenotypic investigation of a New Zealand cohort of childhood-onset retinal dystrophy
Funding information: Cure Kids, New Zealand, Grant/Award Number: 3584; Ombler Charitable Trust, Grant/Award Number: 3626039; Retina New Zealand, Grant/Award Number: 3625913; Save Sight Society of New Zealand, Grant/Award Number: 3625915
Abstract
Inherited retinal diseases are clinically heterogeneous and are associated with nearly 300 different genes. In this retrospective, observational study of a consecutive cohort of 159 patients (134 families) with childhood-onset (<16 years of age) retinal dystrophy, molecular investigations, and in-depth phenotyping were performed to determine key clinical and molecular characteristics. The most common ocular phenotype was rod-cone dystrophy in 40 patients. Leber Congenital Amaurosis, the most severe form of retinal dystrophy, was present in 10 patients, and early onset severe retinal dystrophy in 22 patients. Analysis has so far identified 131 pathogenic or likely pathogenic variants including 22 novel variants. Molecular diagnosis was achieved in 112 of 134 families (83.6%) by NGS gene panel investigation in 60 families, Sanger sequencing in 27 families, and Asper microarray in 25 families. An additional nine variants of uncertain significance were also found including three novel variants. Variants in 36 genes have been identified with the most common being ABCA4 retinopathy in 36 families. Five sporadic retinal dystrophy patients were found to have variants in dominant and X-linked genes (CRX, RHO, RP2, and RPGR) resulting in more accurate genetic counseling of inheritance for these families. Variants in syndromic associated genes including ALMS1, SDCCAG8, and PPT1 were identified in eight families enabling directed systemic care.
1 INTRODUCTION
Retinal dystrophies are a diverse group of inherited disorders of photoreceptors in which developmental and/or degenerative processes lead to visual impairment. The prevalence is approximately 1 in 3500 with the majority of affected patients having a rod-cone dystrophy (RCD) (Bertelsen, Jensen, Bregnhøj, & Rosenberg, 2014; Bocquet et al., 2013). Syndromic associations are found in 25% of cases the most common being Usher syndrome in which RCD and sensorineural hearing impairment coincide (Bocquet et al., 2013). Prevalence and genetic etiology vary geographically with more isolated or consanguineous communities demonstrating a greater burden of disease (Sherwin, Hewitt, Ruddle, & Mackey, 2008). At the severe end of the spectrum is Leber Congenital Amaurosis (LCA) characterized by onset at birth or in infancy, nystagmus, an absence of photoreceptor responses on electroretinogram and severe visual impairment of 20/400 Snellen (1.3 logMAR) to light perception only (Cremers, van den Hurk, & den Hollander, 2002).
Since the initial discovery of RHO causing dominant retinitis pigmentosa in 1990, nearly 300 genes have been associated with retinal dystrophy (Dryja et al., 1990; Wang et al., 2013) (RetNet, https://sph.uth.edu/RetNet/). Many exhibit phenotypic heterogeneity having been associated with more than one retinal disorder. For instance, CRB1 and CRX have been associated with LCA, RCD, cone-rod dystrophy (CORD), and macular dystrophy (MD) (Henderson et al., 2011; Hull et al., 2014; Tsang et al., 2014). Increasingly, with the advent of next generation sequencing, the association of non-syndromic retinal dystrophy with mutations in syndromic genes has been reported including CEP290, CLN3, ALMS1, and BBS1 (Aldrees et al., 2019; den Hollander et al., 2006; Estrada-Cuzcano et al., 2012; Wang et al., 2014). This clinical and molecular heterogeneity presents a diagnostic challenge.
In this study, all patients with childhood onset (less than 16 years of age) disease were ascertained from the New Zealand Database of Inherited Retinal Diseases and extensively phenotyped. Molecular investigations evolved with time from direct Sanger sequencing of candidate genes and genotyping microarrays of specific variants, to next generation sequencing using large gene panels achieving a high diagnostic rate.
2 METHODS
2.1 Clinical assessment
The study protocol adhered to the tenets of the Declaration of Helsinki and received approval from the national and local ethics committees namely the Ministry of Health (NTX/08/12/123) and Auckland District Health Board (A + 4,290). This was a retrospective case series of all patients with childhood onset disease recruited in to the New Zealand database of Inherited Retinal Disease. Written, informed consent was obtained from all participants prior to their inclusion in this study, with parental written consent provided on behalf of the children involved in this study.
Each affected individual underwent a full clinical examination including visual acuity and dilated fundus examination. Where possible, retinal imaging was obtained by conventional 35° fundus color photographs (Topcon Great Britain Ltd, Berkshire, UK), ultra-widefield confocal scanning laser imaging (Optos plc, Dunfermline, UK), 30° or 55° fundus autofluorescence (FAF) imaging (Spectralis, Heidelberg Engineering Ltd, Heidelberg, Germany), and spectral domain optical coherence tomography (OCT) scans (Spectralis). RetCam imaging (Clarity Medical Systems, Inc, CA), was used for examination under general anesthesia using a corneal contact probe with a field of approximately 130°. Full field electroretinography was performed using gold foil electrodes to incorporate the ISCEV standards but in infants and young children skin electrodes were used with modified protocols (Holder & Robson, 2006; McCulloch et al., 2015).
2.2 Molecular investigations
Genomic DNA was isolated from peripheral blood lymphocytes using standard procedures. Arrayed primer extension (APEX) microarray (Asper Biotech Ltd., Tartu, Estonia), was used initially from 2009 to 2016 on 59 patients using a genotyping microarray containing disease causing variants and common polymorphisms. For LCA this comprised >300 variants for eight retinal dystrophy genes (AIPL1, CRB1, CRX, GUCY2D, RPE65, RPGRIP1, LRAT, and MERTK) with an autosomal recessive RCD screen containing >700 disease causing variants for 28 retinal dystrophy genes (ABCA4, AIPL1, CERKL, CNGA1, CNGA3, CNGB3, CRB1, EYS, GRK1, IMPG2, LRAT, MERTK, PDE6A, PDE6B, NR2E3, PROM1, RBP3, RDH12, RGR, RHO, RLBP1, RP1, RPE65, SAG, TULP1, CLRN1, and USH2A) (van Huet et al., 2015; Zernant et al., 2005).
A number of different next generation sequencing (NGS) gene panels were used in this study. Most frequent was the MVL vision panel in 48 probands which in version 2 covers the coding regions of 581 genes associated with inherited ocular disease of which 281 cover isolated and syndromic retinal dystrophy (Molecular Vision Laboratory, Hillsboro, OR). Briefly, NGS libraries were prepared using the Nextera DNA Library Preparation Kit (Illumina, San Diego, CA, USA), analyzed using the DNA 1000 Assay on the Bioanalyzer 2,100 (Agilent Technologies, Santa Clara, CA), and sequenced on Illumina HiSeq 2,500. Reads were aligned to the GRCh37 (hg19) human reference sequence using NextGENe by SoftGenetics, LLC (State College, PA). NGS copy number variant (CNV) analysis was also performed and mutations confirmed by TaqMan qPCR. Array comparative genomic hybridization (CGH) analysis was performed in a few cases using the CytoSure Eye Disease Research array v2 (Oxford Gene Technology [OGT], Begbroke, UK). Array data were analyzed using CytoSure Interpret Software (OGT). Additional panels used included the macular dystrophy panel of 10–14 genes (20 probands), the 14 gene congenital stationary night blindness (CSNB) panel (two probands), and the 6 gene achromatopsia panel (two families).
A panel with 105 genes was performed on four probands' DNA at the Manchester Centre for Genomic Medicine (Manchester, UK) as previously described (O'Sullivan et al., 2012). One proband underwent NGS panel testing of 266 genes at Blueprint Genetics Retinal Dystrophy Panel (Espoo, Finland) (Akinrinade et al., 2015).
Bi-directional Sanger sequencing was performed in probands and available family members where possible. DNA was amplified using specifically designed primers by polymerase chain reaction (PCR) and the resulting fragments sequenced using standard protocols. Sanger sequencing was used for targeted sequencing in 28 probands including 10 of 12 X-linked retinoschisis probands and 6 of 8 X-linked RCD probands.
Variants were identified as novel if not previously reported in the literature and if absent from dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/); NHLBI GO Exome Sequencing Project (ESP), Seattle, WA (http://evs.gs.washington.edu/EVS/); 1,000 genomes project (http://www.1000genomes.org/); the Exome Aggregation Consortium (ExAC) Cambridge, MA (http://exac.broadinstitute.org); and the Genome aggregation Database (gnomAD, http://gnomad.broadinstitute.org) (Abecasis et al., 2010). The likely pathogenicity of novel missense variants was assessed using the predictive algorithms of “Sorting Intolerant from Tolerant” (SIFT, http://sift.jcvi.org) and Polymorphism Phenotyping v2 (PolyPhen-2, http://genetics.bwh.harvard.edu/pph2) (Adzhubei et al., 2010; Kumar, Henikoff, & Ng, 2009). Where relevant, potential splice site disruption was assessed using Splice Site Prediction by Neural Network (https://www.fruitfly.org/seq_tools/splice.html) (Reese, Eeckman, Kulp, & Haussler, 1997). The pathogenicity of novel variants was graded according to the American College of Medical Genetics (ACMG) guidelines to be (P) pathogenic, (LP) likely pathogenic, or (VUS) variant of uncertain significance (Richards et al., 2015). Specifically, the classification criteria of pathogenic or likely pathogenic variants was used in which each criterion was weighted as very strong (PVS1), strong (PS1–4), moderate (PM1–6), or supporting (PP1–5) with scoring rules followed for combining criterion to determine P, LP, or VUS (details in Supplementary Tables 1 and 2).
Mutation nomenclature was assigned in accordance with GenBank Accession numbers with nucleotide position 1 corresponding to the A of the ATG initiation codon with alignment and variant calling using GRCh37 reference genome. Variant nomenclature was assigned according to the Human Genome Variation Society (HGVS) guidelines (http://www.varnomen.hgvs.org) (den Dunnen et al., 2016).
3 RESULTS
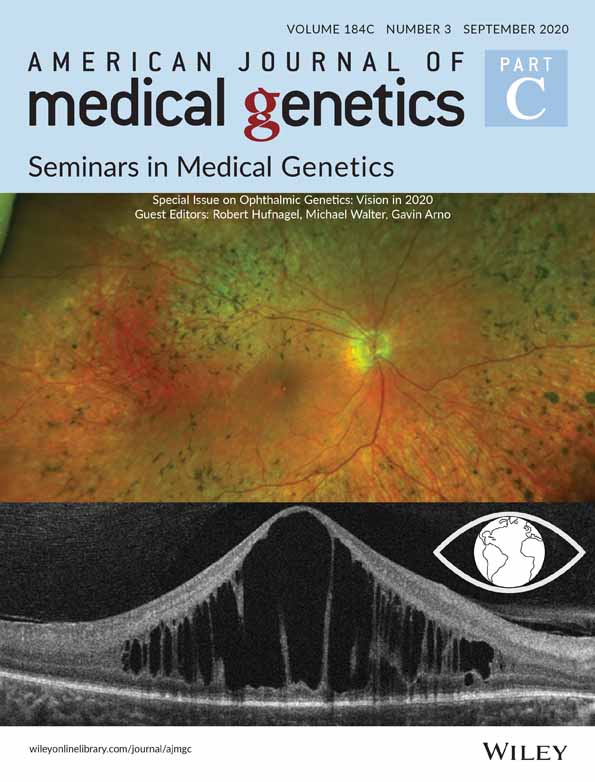
In total, 159 patients (91 male, 68 female) from 134 families have been investigated with 90 Caucasian, 11 Māori, 11 Indian, 9 Asian, 7 Middle Eastern, and 6 Pasifika. Patients presented with visual symptoms at an average age of 60.4 months (median 50 months, range 0–180) with 37 patients presenting in infancy (6 months or less). Fifteen patients presented after failing the pre-school vision screening testing which in New Zealand is performed at the age of 4 years. Patients were reviewed for an average 5.6 years (median 4.8, range 0–22.3). The principal retinal diagnosis was RCD in 40 patients, CORD in 26 patients, early onset severe retinal dystrophy (EOSRD) in 22, MD in 22, X-linked retinoschisis (XLR) in 15, CSNB in 11, LCA in 10, achromatopsia in 6, cone dystrophy with supernormal rod response (CDSR) in 4, and cone dystrophy in 3 (Table 1, Figure 1). The four patients with CDSR are from two families both segregating pseudo-dominant disease and have been previously reported (Kiray, Rapata, Sharp, & Vincent, 2020).
| Diagnosis | Mean age at presentation (median, range) | Mean age at last visit (median, range) | logMAR VA at last review RE (Snellen) | logMAR VA at last review LE (Snellen) | Nystagmus |
|---|---|---|---|---|---|
| RCD (n = 40) | 70.3 months (57, 3–144) |
18.5 years (15, 7–80) |
0.5 (20/63) |
0.5 (20/63) |
0 of 30 |
| CORD (n = 26) | 97.9 months (96, 6–180) |
35.7 years (35, 7–69) |
1.8 (20/900) |
1.6 (20/800) |
2 of 26 |
| EOSRD (n = 22) | 13 months (11, 0–48) |
20 years (12, 2–56) |
3 (HM) |
2 (CF) |
19 of 22 |
| MD (n = 22) | 106.6 months (114, 36–180) |
23.2 years (16, 7–62) |
0.8 (20/125) |
0.9 (20/160) |
0 of 12 |
| XLR (n = 15) | 68 months (60, 24–132) |
22.3 years (11, 7–57) |
1.1 (20/250) |
0.5 (20/63) |
0 of 14 |
| CSNB (n = 11) | 19 months (6, 0–60) |
23 years (16, 2–62) |
0.7 (20/100) |
0.7 (20/100) |
7 of 11 |
| LCA (n = 10) | 2 months (2, 0–6) |
6 years (6, 0–9) |
4 (PL) |
4 (PL) |
10 of 10 |
| Achrom (n = 6) | 1 month (0, 0–2) |
23 years (21, 4–48) |
1 (20/200) |
1 (20/200) |
6 of 6 |
| CDSR (n = 4) | 9 months (6, 0–24) |
27.5 years (27.5, 3–52) |
0.9 (20/160) |
1.0 (20/200) |
2 of 4 |
| CD (n = 3) | 93 months (132, 3–144) |
14 years (15, 7–20) |
0.9 (20/160) |
1 (20/200) |
0 of 2 |
- Abbreviations: Achrom, achromatopsia; CD, cone dystrophy; CDSR, cone dystrophy with supernormal rod response; CF, counting fingers; CORD, cone rod retinal dystrophy; CSNB, congenital stationery night blindness; EOSRD, early onset severe retinal dystrophy; HM, hand movements; LCA, Leber congenital amaurosis; LE, left eye; MD, macular dystrophy; PL, perception of light; RCD, rod-cone dystrophy; RE, right eye; VA, visual acuity; XLR, X-linked retinoschisis.
All 10 patients with LCA presented with severe visual impairment, nystagmus, and extinguished or barely recordable ERG. Four had oculodigital reflex and three had severe circumscribed macular chorioretinal atrophy at presentation. Two had developmental delay. The six patients with achromatopsia similarly presented with nystagmus and visual impairment from birth or early infancy but at last review average 23 years of age had an average visual acuity of 1.0 logMAR (Snellen 20/200). Electrophysiology performed in all patients identified normal rod function with an absence of responses on bright flash and 30 Hz flicker indicating a lack of cone function.
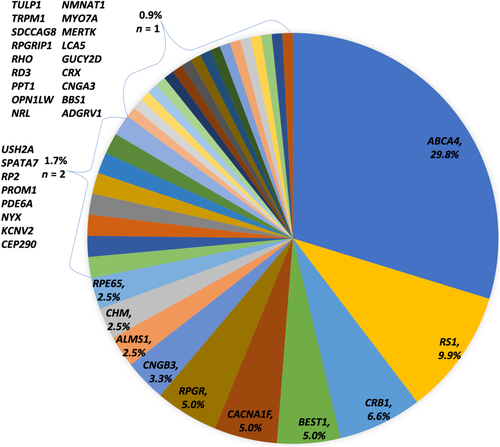
Overall 112 of 134 families received a probable molecular diagnosis, a positive rate of 83.6%. There were an additional nine families who had at least one VUS but with their associated phenotype making the diagnosis probable. In total, 140 variants were found in 36 genes including 25 novel variants in 17 different genes (Figure 2, Table 2, full list of variants, and associated references in supplementary Table 3). Classification of these novel variants according to the ACMG found 13 were predicted to be pathogenic, 9 likely pathogenic, and 3 as VUS due to mixed in silico predictive outcomes and/or a lack of available segregation (Richards et al., 2015). NGS gene panel testing solved 60 families, Sanger sequencing of candidate genes 27 families, and Asper microarray 25 families. Of the 60 families solved by NGS gene panel testing, 29 had undergone initial Asper microarray testing which was negative in 25 probands and found single variants in 4 probands later found to have recessive disease. Of the 42 probands who underwent the 281 MVL gene panel, 34 were solved (81.0%).
| Gene, GenBank Accession Number | Phenotype(s) | Novel variant | ||||
|---|---|---|---|---|---|---|
| Nucleotide | Protein | Polyphen2 | Sift | ACMG score | ||
| ABCA4, NM_000350.2 | CORD | c.4225A > G | p.(Ile1409Val) | 0.003 B | 0.014 D | VUS |
| CD | c.4288_4294del | p.(Leu1430Thrfs*4) | – | – | P | |
| MD | c.5584G > T | p.(Gly1862Cys) | 1.000 PD | 0.001 D | LP | |
| CRB1, NM_201253.2 | EOSRD | c.1631 T > C | p.(Leu544Ser) | 0.999 PD | 0.001 D | VUS |
| BEST1, NM_004183.3 | MD | c.94C > G | p.(Leu32Val) | 1.000 PD | 0.214 T | VUS |
| MD | c.892 T > G | p.(Phe298Val) | 1.000 PD | 0.000 D | LP | |
| CACNA1F, NM_001256789.3 | CSNB | c.2649dupC | p.(Ser884Glnfs*8) | – | – | P |
| CSNB | c.4622-2A > T | p.(?) | – | – | P | |
| RPGR, NM_001034853 | RCD | c.550C > T | p.(Gln184*) | – | – | P |
| RCD | c.597 T > G | p.(Tyr199*) | – | – | P | |
| RCD | c.1379delT | p.(Leu460Tyrfs*16) | – | – | P | |
| CNGB3, NM_019098.4 | Achromatopsia | c.991-3delTAG | p.(?) | – | – | LP |
| CHM, NM_000390.4 | Choroideremia Choroideremia |
c.525delA | p.(Glu177Cysfs*20) | – | – | P |
| c.1637_1649del | p.(Leu547Thrfs*5) | – | – | P | ||
RP2, NM_06915.3 |
RCD | c.945_946insT | p.(Asn316*) | – | – | P |
| SPATA7, NM_018418.5 | RCD | c.1028G > A | p.(Arg343Lys) | 0.998 PD | 0.022 D | LP |
| CNGA3, NM_001298.3 | Achromatopsia | c.624C > G | p.(Tyr208*) | – | – | LP |
| CRX, NM_000554.6 | LCA | c.534delG | p.(Leu179Trpfs*8) | – | – | P |
| LCA5, NM_001122769.3 | LCA | c.194delC | p.(Pro65Leufs*46) | – | – | LP |
| MY07A, NM_000260.4 | Usher syndrome | c.5916G > A | p.(Trp1972*) | – | – | LP |
| NRL, NM_006177.5 | RCD | c.152C > G | p.(Pro51Arg) | 1.000 PD | 0.017 D | LP |
| RD3, NM_001164688.1 | LCA | c.127C > T | p.(Gln43*) | – | – | P |
| RPGRIP1, NM_020366.3 | LCA | c.711delA | p.(Lys239Serfs*36) | – | – | P |
| c.832delC | p.(Arg278Aspfs*15) | – | – | P | ||
| TULP1, NM_003322.6 | EOSRD | c.1382 T > C | p.(Leu461Pro) | 1.000 PD | 0.000 D | LP |
- Note: Polyphen2 score < 0.90 benign (B), ≥0.95 probably damaging (PD). Sift score < 0.05 deleterious (D), >0.05 tolerated (T).
- Abbreviations: ACMG, American College of Medical Genetics; BCM, blue cone monochromatism; CD, cone dystrophy; CDSR, cone dystrophy with supernormal rod response; CORD, cone rod retinal dystrophy; CSNB, congenital stationery night blindness; EOSRD, early onset severe retinal dystrophy; LCA, Leber congenital amaurosis; MD, macular dystrophy; NCL, neuronal ceroid lipofuscinosis; P, pathogenic; PL, likely pathogenic; RCD, rod cone dystrophy; SLS, Senior-Løken syndrome; VUS, variant of uncertain significance; XLR, X-linked retinoschisis.
The most frequently identified gene was ABCA4 affecting 40 patients from 36 families with 23 patients manifesting maculopathy with cone and rod dysfunction, isolated macular dystrophy in 14 patients, maculopathy with cone dysfunction in 2 patients and EOSRD in 1 patient. Fifteen male patients from 12 families had X-linked retinoschisis characterized by macular schises, electronegative ERG, and myopia. Probable causative variants were found in 10 families by Sanger sequencing of RS1, the remaining two solved by MVL gene panel investigation. CRB1 related disease was found in 11 patients from eight families manifesting with severe early onset disease in nine patients and RCD in two patients. Although three patients had onset at birth, they described having navigational vision until at least their late 20s and were, therefore, classed as EOSRD not LCA. Refractive data available in eight patients indicated hyperopia average + 5.5 D spherical equivalent. CACNA1F related CSNB was found in eight patients from six families including a family with an extended pedigree of severely affected males and females that has been previously reported (Hope et al., 2005). The female included in this study had onset of nystagmus and reduced vision from birth and at last review age 48 years had vision of 0.9 logMAR each eye (Snellen 20/160).
There were 56 families with sporadic presentations in whom there was no family history of disease or consanguinity to indicate specific inheritance patterns and the clinical phenotype was not indicative of an inheritance pattern. Variants were found in 50 families consistent with recessive disease in 46, dominant disease in 2, and X-linked disease in 2. Dominant variants included a novel premature truncating variant in CRX, c.534delG (p.Leu179Trpfs*8) confirmed to be de novo in the proband who had presented with LCA; and a previously reported heterozygous variant in RHO, c.403C > T:p.(Arg135Trp) in a proband with RCD presenting age 4 years (Sung et al., 1991). This latter proband has unaffected, non-consanguineous parents and three unaffected siblings with segregation pending. De novo and intrafamilial phenotypic variability of this variant has been reported (Abdulridha-Aboud, Kjellström, Andréasson, & Ponjavic, 2016). The first sporadic patient found to have X-linked disease, was an only child of non-consanguineous Vietnamese parents. He presented at age 2 with nystagmus and nyctalopia and was found to have EOSRD with no rod or cone mediated function detectable on ERG age 3 years. At last review age 16 years, visual acuity was 1.3 logMAR (20/400 Snellen) with fine vertical nystagmus, high myopia (spherical equivalent −7.5D each eye), disc pallor, and peripheral granular pigmentation. MVL gene panel found a previously reported variant in RP2, c.413A > G (p.Glu138Gly), that is absent from gnomAD (Miano et al., 2001). In addition, a homozygous variant in GUCY2D was found, c.343 T > C:p.(Ser115Pro) which has an allele frequency of 1 in 200 East Asians in gnomAD and has been reported as likely benign (Huang et al., 2013). Unfortunately, maternal examination and segregation has not been possible. The second sporadic proband found to have X-linked disease presented with poor vision and nyctalopia age 5 and at last review age 7 had vision of 0.22 (20/32) right eye and 0.30 (20/40) left eye. He was found by MVL 281 gene panel to have a novel hemizygous variant in RPGR c.550 C > T:p.(Gln184*) which is absent in gnomAD. The variant was confirmed in his unaffected mother.
The molecular results helped to redefine clinical diagnoses in three probands. A 3 month old boy of non-consanguineous unaffected parents with two older unaffected siblings, presented with nystagmus and poor fixation. ERG performed with lid electrodes at 3 months of age found no rod response and reduced cone response and a diagnosis of likely EOSRD was given. At last review age 18 months old, visual acuity with both eyes open was 0.7 logMAR (20/100 Snellen) with myopic refraction of −2 D each eye and a normal fundus. MVL 281 gene panel testing identified two variants in TRPM1 namely c.296 T > C:p.(Leu99Pro), previously reported, and c.3067G > A:p.(Ala1023Thr), not previously reported and present in 1 allele of 249,096 on gnomAD with parental DNA unavailable for segregation (Audo et al., 2009). On review of phenotype, a diagnosis of CSNB has been made. Two probands presenting with EOSRD were found to have probable disease causing variants in ALMS1 leading to pediatric assessment and sub-specialty investigation and diagnosis of Alström disease.
A potential non-syndromic association with ALMS1 was found in a Samoan patient who developed decreased central vision age 10 years. At age 14 vision was counting fingers each eye with marked bilateral macular atrophy. Apart from an increased body mass, the patient had no hearing reduction, diabetes, or other systemic problems. EDTs found undetectable pattern ERG, mild reduction in scotopic responses, and absent photopic bright flash consistent with a moderately severe CORD. NGS 281 gene panel investigation found a homozygous missense variant in ALMS1 c.571C > A:p.(Leu191Met), gnomAD allele frequency 9.64e−5. The variant was predicted to be pathogenic by Polyphen2 (score 0.95) but tolerated by Sift (0.135) and has been classified as a VUS.
A further three patients initially presented with retinal dystrophy and were subsequently diagnosed with syndromic disease that was then confirmed on sequencing; neuronal ceroid lipofuscinosis with EOSRD due to variants in PPT1, Senior-Løken syndrome with RCD due to variants in SDCCAG8 and Bardet Biedl Syndrome with CORD due to a homozygous variant in BBS1. The patient with PPT1 variants identified on MVL gene panel testing, presented with visual disturbance age 4 years with neurological decline only evident from age 10 years. The 44 year old patient with Bardet Biedl syndrome and CORD presented at 10 years of age with decreased color vision and dyslexia. She had a history of polydactyly, brachydactyly, increased body mass index, and menstrual irregularities. Mild renal impairment of unknown cause had been diagnosed in her 20s. MVL gene panel investigation found a previously reported homozygous variant in BBS1 c.1169 T > G:p.(Met390Arg) (Mykytyn et al., 2002). The patient with Senior-Løken syndrome initially presented with RCD but shortly after was admitted in end stage renal failure at age 11 years prior to molecular diagnosis of compound heterozygous variants in SDCCAG8 (Tay & Vincent, 2020). Three families presented with sensorineural hearing loss and signs of RCD; all were confirmed to have Usher syndrome with variants found in USH2A, ADGRV1, and MY07A.
Of the 19 homozygous variants identified, consanguinity was known in seven families. Four of six Pasifika families with no known consanguinity were found to have homozygous variants (in CRB1, RD3, SPATA7, and ALMS1). Parents of each proband were from the same island specifically Rarotonga, Tonga, Samoa, and Samoa, respectively. Enrichment of variants in isolated populations has been described (Avela et al., 2019; Sherwin et al., 2008). Considering non homozygous variants, recurrent alleles found in this study largely arose within ABCA4 (Supplementary Table 3). Six Caucasian probands carried p.(Gly863Ala), six Caucasian probands carried p.(Cys1490Tyr), and three probands (two Caucasian, one Indian) carried p.(Pro1380Leu).
Of the 13 unsolved families, two probands with a phenotype suggestive of North Carolina Macular Dystrophy had negative Sanger sequencing of candidate genes and are undergoing whole-genome sequencing investigation; three probands had negative Asper microarray testing and are awaiting further gene panel testing; and eight probands have undergone negative gene panel testing, and whole-exome sequencing is now planned. These eight families include a patient with LCA and macular chorioretinal atrophy; siblings with high myopia, macular atrophy, cataract, and retinal detachment suggestive of a Knobloch phenotype whose parents are consanguineous; and four probands with sporadic RCD. The seventh proband, a 21 year old male presented at 18 months with nystagmus, head shaking, macular atrophy with peripheral pigmentary change, and severely reduced ERG consistent with an EOSRD and is systemically well. Three variants in VPS13B were identified, c.5681C > T:p.(Thr1894Met) maternal, likely benign, c.6491A > G (p.Asn2164Ser), maternal, likely benign and c.7226C > T:p.(Pro2409Leu), paternal, variant of uncertain significance. In the absence of systemic features of Cohen Disease and with the maternally inherited variants predicted benign, this patient remains unsolved. NGS panel in the eighth proband found a single variant in GUCY2D, c.2302C > T:p.(Arg768Trp) which has been previously reported in recessive GUCY2D related disease (Lotery et al., 2000). Although heterozygous disease has been reported it typically manifests as a cone-rod dystrophy with causative variants clustered in exon 13 and has not been reported with the 768 codon (Kelsell et al., 1998).
4 DISCUSSION
This study has investigated the phenotype and molecular diagnosis of patients from the NZ database of Inherited Retinal Disease presenting in childhood. Of the 159 patients, 32 presented with severe disease and had the earliest age of onset, poorest vision, and most reduced ERG compared with other groups (Table 1). For patients presenting with early onset retinal dystrophy, particularly those with LCA, systemic assessment is warranted to exclude an underling syndromic diagnosis or potential associated syndromic findings. In LCA, patients may have developmental delay and learning difficulties as found in two probands, as well as other associations such as hearing loss, cerebellar vermis hypoplasia, and cardiomyopathy (Falk et al., 2012; Marshall et al., 2007). Syndromic findings may only present later. For instance, Senior-Løken syndrome characterized by nephronophthisis and EOSRD usually presents in the first decade with visual symptoms but renal involvement may not present until the second decade as in the patient with SDCCAG8 related disease in this series (Otto et al., 2005; Stone et al., 2011).
The most frequently identified molecular cause was ABCA4 in 36 families representing 29.8% of diagnoses followed by RS1 in 9.9%, CRB1 in 6.6% and then BEST1, CACNA1F, and RPGR each with 5%. In comparison, a series of 85 consecutive children with childhood onset (16 years or younger) inherited retinal disease investigated by NGS gene panel testing (Manchester 105 or 177 gene panel) found most frequently CACNA1F in 14.9%, ABCA4 11.9% and then CNGB3, NYX, TRPM1, and CNGA3 all with 6% (Taylor et al., 2017). Their population differed in underlying phenotype as 21.2% of cases had CSNB whereas 8.2% of probands had CSNB in this series. They achieved a probable/possible molecular diagnosis of 84.7% similar to our series although their panel investigation represents a single testing point. Thirty families in our series in whom Asper microarray was initially performed subsequently needed NGS gene panel testing to achieve a molecular diagnosis and were investigated over years as the testing methods available evolved. The primary testing method now preferred is the MVL 281 gene panel and the diagnostic rate for this panel alone in our series was 81.0%. This high success rate for molecular diagnosis is comparable to the Manchester series and other studies have also reported a higher success rate in the pediatric population (Shanks et al., 2013).
Recurrent alleles in this study were mainly found in ABCA4. These variants have previously been reported at an increased prevalence in affected patients, but this prevalence varied geographically suggesting potential founder effects (Fujinami et al., 2019). Founder mutations in ocular genes including PDE6B and ADAMTLS4 have previously been reported in Māori and Pasifika families (van Bysterveldt, Al Taie, Ikink, Oliver, & Vincent, 2017; Vincent et al., 2017). Within this specific study no recurrent alleles were found within the nine Māori and Pasifika families manifesting recessive disease as eight different genes were involved and greater numbers of affected families would be needed to identify recurrent alleles. However, four novel variants and two variants not previously reported in disease (but found in general population databases) were identified. It would be interesting to look for enrichment of these alleles within population specific reference genome databases which are not currently available and are the focus of future study.
Molecular diagnosis can help to identify those patients who need specific systemic investigations as found in the probands diagnosed with Alström disease and now under cardiac and metabolic monitoring. For one patient with CORD, NGS identified a homozygous missense VUS in ALMS1 not previously reported in an affected patient but found in gnomAD at a low allele frequency of 9.64e−5. The majority of pathogenic variants in ALMS1 are premature truncating codons clustering in exons 8, 10, and 16; rarely, missense variants have been reported (Marshall et al., 2015). There is significant inter and intra-familial variability in phenotypic expression and non-syndromic associations with variants in ALMS1 have been found (Aldrees et al., 2019; Paisey et al., 2019). It, therefore, remains a possible but unproven cause of this patient's CORD.
Molecular diagnosis also facilitates informed genetic counseling as to risk of further affected children or of passing on the disease. This is particularly useful in situations where there is no family history or consanguinity. Two sporadic probands were found to have dominant disease and two were found to have X-linked disease allowing accurate counseling. For the patient with a novel, de novo CRX variant, this also meant that the risk of the parents having a further affected child was extremely small. Molecular diagnosis can also trigger reassessment of the phenotype as found in the proband presenting with EOSRD but with TRPM1 variants indicating a diagnosis of CSNB which has a better prognosis (Bijveld et al., 2013).
NGS gene panels, provides an unbiased and efficient approach to investigating retinal dystrophy patients as has been found in this study (Audo et al., 2012; Taylor et al., 2017). This is particularly the case when the panel also covers known intronic regions of importance such as the common CEP290 c.2991 + 1655A > G and when CNV analysis is also routinely performed as found in the 281 gene panel frequently used in this study (Eisenberger et al., 2013). For the eight unsolved families who have undergone gene panel testing, there may have been variants missed by poor coverage, noncoding region variants not specifically covered in the panel, or small CNVs not identified. Whole-exome sequencing particularly when performed as a trio with parental sequencing may help solve some of these cases but ultimately whole genome sequencing with its better coverage and read depths may be needed (Nishiguchi et al., 2013).
Gene therapy for RPE65 related disease in the form of an adeno-associated virus vector- based therapy Voretigene Neparvovec (Spark Therapeutics, Philadelphia, PA) is now approved by both the US Food and Drug Administration and the European Medicines Agency. Other genes with therapies trialed or in development include ABCA4, CEP290, CHM, CNGA3, CNGB3, GUCY2D, MERTK, MYO7A, PDE6A, RHO, RLBP1, RPGR, RS1, and USH2A (https://clinicaltrials.gov). These potential therapies encompass 70 families in this series. Achieving a molecular diagnosis not only helps to inform prognosis and inheritability but is an essential prerequisite for trial eligibility.
DISCLOSURE OF INTERESTS
John Pei-Wen Chiang is director of the Molecular Vision Laboratory, Oregon, USA.
Andrea L. Vincent, Sarah Hull, and Gulunay Kiray have no conflicts of interest to declare.
Open Research
DATA AVAILABILITY STATEMENT
Data available in article supplementary material.