Ophthalmic genetics practice and research in India: Vision in 2020
Abstract
Ophthalmic genetics is a much needed and growing area in India. Ethnic diversity, with a high degree of consanguinity, has led to a high prevalence of genetic disorders in the country. As the second most populous country in the world, this naturally results in a significant number of affected people overall. Practice involves coherent association between ophthalmologists, genetic counselor and pediatricians. Eye genetics in India in recent times has witnessed advanced research using cutting edge diagnostics, next generation sequencing (NGS) approaches, stem cell therapies, gene therapy and genomic editing. This article will highlight the studies reporting genetic variations in the country, challenges in practice, and the latest advances in ophthalmic genetic research in India.
1 INTRODUCTION
India is a large and diverse country, with population subsets of different ethnicities. Certain population groups have a high degree of consanguinity, and therefore a higher prevalence of genetic disorders. Here, we delve into the landscape of genetic eye diseases in India, their prevalence, challenges faced in management, and possible solutions. We also discuss the advances in basic sciences research in ophthalmic genetics in India, and a vision for the future. Among the genetic eye conditions seen in India, the more prevalent ones are listed below. While there are many significant articles reporting their underlying genetic variations, our attempt is to make the description as comprehensive as possible, while including genomic databases, mutations, and genotype–phenotype correlation from India.
2 CORNEAL DYSTROPHIES
2.1 Landscape and demographics in India
Genetic diagnosis is increasingly being included into the classification of corneal dystrophies. (Weiss et al., 2015) Genotyping has revealed genotypic and phenotypic heterogeneity among genes causing corneal dystrophies. India sees a large volume of patients affected by this group of diseases, wherein, 8.1% of keratoplasties or corneal transplants in southern India, are performed for corneal dystrophies (Pandrowala, Bansal, Vemuganti, & Rao, 2004).
2.2 Advances in diagnostics: Novel mutations, sequencing studies, and molecular diagnostics
Showing phenotypic heterogeneity, TGFBI gene is associated with multiple distinct dystrophy phenotypes, notably, lattice corneal dystrophy (LCD, Type 1), and granular corneal dystrophy (GCD). In mutational analysis of TGFB1 gene in 80 patients from Northern India, which had been phenotypically diagnosed as LCD and GCD; missense mutations, c.370C>T (p.Arg124Cys) and c.1663C>T (p.Arg555Trp) were found most commonly (Paliwal et al., 2010). Two novel mutations, c.1548C>G (p.Ser516Arg) and c.1675C>G (p.Leu559Val), were identified in patients who were clinically diagnosed with GCD. The patients with novel mutations did not require any surgical intervention so the nature of deposits could not be assessed histopathologically. Further, on analysis of TGFB1 gene in patients diagnosed with Avellino Corneal Dystrophy in North India, the p.Arg141His mutation was reported (Paliwal, Gupta, Tandon, Sharma, & Vajpayee, 2009). The above studies concluded, therefore, that the phenotypic presentation of the disease can be variable and can overlap.
Macular corneal dystrophy (MCD) is characterized by an early onset of central stromal haze gradually extending to the periphery of the cornea. Mutations in the carbohydrate sulfotransferase-6 (CHST6) gene on 16q22 cause MCD. Paliwal et al. did a study on 30 patients affected with MCD in North India. Sequence analysis of CHST6 revealed seven homozygous and five heterozygous changes of which four were novel homozygous missense mutations. The novel p.Ser54Tyr, p.Gln58Arg, p.Leu293Phe, and p.Leu59His substitutions were identified in this study. (Paliwal et al., 2012) Correlation with histopathology was not possible in all the patients, indicating that the mutation-negative cases may represent phenocopies. Absence of pathogenic changes in 13 individuals suggests genetic heterogeneity or changes in the CHST6 regulatory regions. Various studies have reported absence of CHST6 mutations in one or both alleles in MCD individuals.
Fuchs endothelial corneal dystrophy (FECD), is classified based on age of onset as early onset (<40 years) and late onset. Mutations in the COL8A2 gene have been identified in early onset FECD. A small percentage of mutations in SLC4A11, ZEB1 (also known as TCF8), LOXHD1, and AGBL1 genes have been reported in sporadic late-onset FECD cases in the Caucasian, Chinese, African-American, and Northern European ethnic groups. Rao et al. studied 52 cases of late onset FECD in India. Mutations in SLC4A11 contribute to approximately 11% and mutations in LOXHD1 contribute to 2% of the FECD cases in this study. A comparison of the role of ZEB1 across multiple ethnicities shows that the contribution of this gene in our Indian population is concurrent with other reports (Rao et al., 2018). In another study it was found that the single nucleotide polymorphisms (SNPs) of TCF4 gene, rs613872 and rs1759573 were associated with a higher risk of sporadic late-onset FECD in Indian patients (Rao, Tharigopala, Rachapalli, Rajagopal, & Soumittra, 2017).
Congenital hereditary endothelial dystrophy (CHED) has been described to be caused by the bicarbonate transporter-related SLC4A11 gene. A study from North India reported an unusual case of a 45-year-old female presenting with CHED which was confirmed on histopathology. Analysis of the SLC4A11 gene showed a novel variant c.1244G>A (p.Ser415Asn), and mutation c.1156T>C (p.Cys386Arg), which were present in the heterozygous state. The authors concluded that heterozygous mutations may have a mild effect on the protein function and levels and thereby may have resulted in the late onset of the disease (Kumawat et al., 2016). Further, four novel mutations, c.785C>T (p.Thr262Ile), c.1831T>C (p.Cys611Arg), c.1249G>A (p.Gly417Arg), and c.2170C>G (p.His724Asp) in SLC4A11 gene have been found in a study from six Indian families (Kodaganur et al., 2013). Recently, it has been found that heterozygosity for SLC4A11 mutations in the parents of children with autosomal recessive CHED appears to be a risk factor for the development of FECD (Chaurasia, Ramappa, Annapurna, & Kannabiran, 2020).
Keratoconus is a bilateral, often asymmetric noninflammatory ectatic corneal disorder which may be idiopathic in origin, may have a genetic association and hereditary natures and may or may not be associated with ocular or systemic comorbidities or risk factors. It is not included conventionally in the list of corneal dystrophies and is considered a separate entity. However, it merits a mention in this segment as per newer knowledge and understanding. A genetic basis for keratoconus has been increasingly established. LOX, ZNF469 (Brittle cornea syndrome), TGFB1 (multiple corneal epithelial–stromal dystrophies), VSX1 (visual system homebox1), and COL5A1 are the genes which have been associated with keratoconus (Bykhovskaya, Margines, & Rabinowitz, 2016). Further the role of microRNAs (miRNAs) in the pathogenesis of keratoconus has been studied wherein mutations in MIR184 were found to be responsible for familial keratoconus (Hughes et al., 2011). Among Indian patients, c.525G>C (Gln175His), one novel mutation in VSX1 gene, predicted to cause a pathogenic change was identified in a study of 66 patients affected with keratoconus (Paliwal, Singh, Tandon, Titiyal, & Sharma, 2009). This substantiates the importance of the VSX1 gene but its precise role in disease causation needs further investigation.
This knowledge of mutations and/or variations involved in corneal dystrophies has helped in disease identification when the diagnosis cannot be confirmed clinically, which in turn aids in prognosticating, as well as providing genetic counseling to families and their relatives. The logical next step to sequencing studies, would be to develop novel treatment strategies for corneal dystrophies based on this knowledge. Corneal organoid research, and gene therapy in India, has laid initial ground for this, and will be discussed later in this article (Susaimanickam et al., 2017).
3 GENETICS OF INHERITED RETINAL DYSTROPHIESAND COMPLEX MULTIFACTORIAL RETINAL DISEASES
3.1 Landscape and demographics of inherited retinal dystrophies in India
3.1.1 Prevalence of inherited retinal dystrophies in population
While the global prevalence of retinitis pigmentosa (RP) is estimated to be 1 in 4,000, in India it is found to be much higher (Hamel, 2006). One of the largest surveys conducted in India to find the prevalence of RP s reported that 1 in 372 in rural population, and 1 in 930 in urban populations are affected by RP (Sen et al., 2008). This study was conducted by doing a fundus examination of 7,461 individuals among the general population, above the age of 40 years, of which 13 patients were found to have RP. Further, a population based, cross-sectional study from Central India, assessed the fundus photo of 4,543 individuals (Nangia, Jonas, Kulkarni, & Matin, 2011). They found that 1 in 750 individuals in rural Central India have RP. Extrapolating this to the total Indian population, it can be estimated that half a million individuals have RP in India, and 1.4 million carry the genetic mutations for RP. In another study from South India, a complete eye examination of 2,513 patients attending the out-patient services of a tertiary eye hospital, 673 patients were found to have a genetic eye disease. RP was present in 430 of these 673 patients with most patients having a high degree of consanguinity and autosomal recessive pattern of inheritance (Kumaramanickavel, Joseph, Vidhya, Arokiasamy, & Shetty, 2002).
3.2 Genomic sequencing of inherited retinal dystrophies patients
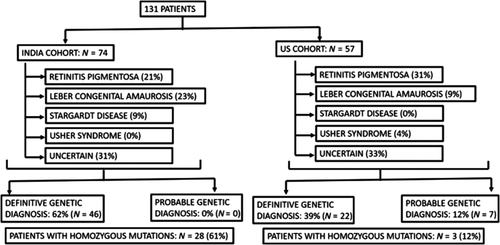
Next generation sequencing (NGS) approaches to determine causative mutations in patients with inherited retinal dystrophies (IRDs) have been steadily growing in India over the last decade. It has helped researchers study the prevalence of causative mutations in India, which are different when compared to data from the West. Among the most significant, is a study reporting comparative data of causative mutations in patients from India and the United States (US) (Yohe et al., 2020). In this study, NGS for targeted retina panel was done for 74 Indian and 57 American patients with retinal dystrophies. The occurrence of different types of retinal dystrophies were found to be different, as well as the genes and mutations involved. Overall RP, Leber's congenital amaurosis (LCA), and Stargardt's disease were the most common IRDs in both cohorts. Homozygous mutations were more prevalent in the Indian cohort, 61%, as compared with 12% in the US cohort. This further signifies the high degree of consanguinity in the Indian population. Also, the prevalence of certain mutations in genes, for example, CEP290 gene in LCA, seen more commonly in the US population, was not found to be so in the Indian cohort, where GUCY2D gene mutations were found to be more common. Only USH2A and CERKL genes were found to be mutated in both cohorts (Figure 1).
Homozygosity mapping has been used extensively to find novel mutations in Indian population, especially in patients with autosomal recessive retinitis pigmentosa (arRP). It provides a rapid method of assessment, especially where consanguinity is prevalent. Specifically, the genes TULP1, RLBP1, ABCA4, RPE65, and RP1 were studied using homozygosity mapping. Two frame-shift, two missense, and one nonsense mutation was found, of which four had never been reported previously (Singh, Jalali, Narayanan, & Kannabiran, 2009). Another study reported novel IRD mutations using homozygosity mapping guided NGS; RDH12 c.832A>C (p.Ser278Arg) mutation in LCA, ABCA4 c.1462G>T (p.Glu488Ter) mutation in Cone-Rod Dystrophy, and CDHR1 c.1384_1392delCTCCTGGACinsG (p.Leu462AspfsTer1) mutation in arRP families. The phenotype of these mutations correlated with previously reported different mutations in the same gene (Sundaramurthy et al., 2016).
In among the only studies reporting mutation analysis in Stargardt's Diseases in India, the following candidate Stargardt related genes were studied (ABCA4, ELOVL4, CNGB3, PROM1, PRPH2, and CRB1). This case series of 5 patients found mutations in only the ABCA4 gene, 2 of which were novel (Battu et al., 2015). In a study on 30 families with Bardet-Biedl syndrome (BBS) in India, 22 families reported causative mutation in known Bardet-Biedl syndrome genes. In 2 families, mutation was found in the ciliopathy related ALMS1 gene. Mutations in the ARL6 gene, among which c.272T>C (p.Ile91Thr) was novel, were responsible for 18% of the identified variations (Sathya Priya et al., 2015). In Indian patients with LCA, CEP290 gene mutations have been found to be remarkably low as compared to European and American literature. Mutations in GUCY2D gene on the contrary have been reported to be in higher frequency (Srikrupa et al., 2018). Congenital stationary night blindness (CSNB) patients seen in India, were screened for LRIT3, TRPM1, GRM6, GPR179, and NYX genes. Among the eight families screened, TRPM1 gene was found to be majorly associated with CSNB (Malaichamy et al., 2014). Choroideremia, an X-linked inherited dystrophy, is caused by mutations in the CHM gene. Patients from India have been reported to have c.820-1G>C and c.653G>C (p.Ser218X) mutations (Battu et al., 2016).
3.3 Analysis of IRD from genomic database
In silico research on publically available genome databases from South Asia, for RP revealed some interesting observations (Hariprakash et al., 2018). This study compiled six datasets with 1,213 human genome and exome sequences. The curated database, called South Asian Genomes and Exomes (SAGE) enlists various conditions, and the authors of the curated database, published RP statistics as an example. Their analysis showed presence of autosomal recessive disorders more commonly in India.
3.4 Genetic studies of complex multifactorial retinal diseases from India
While genetics of multifactorial retinal diseases is a vast topic, in this section we present significant studies from the Indian context, mostly reporting genetic polymorphisms seen in the Indian population.
3.4.1 Age-related macular degeneration
The India age-related eye disease study (INDEYE), reported single nucleotide polymorphisms (SNPs) in both dry or nonneovascular age-related macular degeneration (AMD) and neovascular AMD or nAMD. Two variants in ARMS2/HTRA1 gene [rs10490924, c.205G>T (p.Ala69Ser) and rs2672598, -487T>C] were associated with an increased risk of early AMD. No association was found with CFH Tyr402His variant [rs1061170, c.1277T>C (p.Tyr402His)], C2 (rs547154, G>T), or CFB (rs438999, A>G) (Sundaresan et al., 2012). In nAMD, CFH Tyr402His variant [rs1061170, c.1277T>C (p.Tyr402His)], ARMS2 [rs10490924, c.205G>T (p.Ala69Ser)], C2 (rs547154, G>T), ABCA1 (rs1883025, C>T), and VEGFA (rs4711751, T>C) were found to be associated with an increased risk. No association of TLR3 (rs3775291, C>G/C>T), FRK (rs1999930, C>A/C>G/C>T), CFD (rs3826945, C>G/C>T), and LIPC (rs10468017, C>T) was found with nAMD (Rajendran et al., 2018).
3.4.2 Diabetic retinopathy
Association of SNPs in seven identified genes, was studied in Indian patients with type 2 diabetes mellitus (DM) and diabetic retinopathy (DR). The genes studied were AGER, ICAM, PEDF, EPO, AKR1B1, HTRA1, and HFE. The study confirmed a significant association of one polymorphism (rs2070600, C>T in AGER) (Balasubbu et al., 2010). Further, in another study in proliferative diabetic retinopathy (PDR) in Indian patients with Type 2 DM, gene polymorphisms in the 5′ UTR and promoter region of VEGF were reported to increase risk of DR (Suganthalakshmi et al., 2006). The mitoscriptome analysis in Indian cadaveric eyes with DR reported expression of genes responsible for angiogenesis, antioxidant defense mechanism, ATP production and apoptosis. Specifically, the CASP8 gene was found to be highly regulated (Govindarajan, Mathews, Srinivasan, Ramasamy, & Periasamy, 2017).
3.4.3 Myopia
The membrane frizzled related protein, MFRP gene SNPs have been reported to be associated with high axial length. Among the Indian population, in MFRP gene variant rs36015759 [c.492C>T (pTyr164=)] was reported more commonly in the control population. It was, therefore, inferred that this SNP in MFRP, was associated with a significantly low risk for myopia (Sharmila et al., 2014).
4 HEREDITARY OPTIC NEUROPATHIES—GENETIC RESEARCH FROM INDIA
4.1 Leber's hereditary optic neuropathy
Hereditary optic neuropathies constitute approximately 10% of patients presenting with optic neuropathy without disc edema, as observed in a study from North India from a tertiary care neuro-ophthalmology clinic (Dhiman et al., 2018). The most common hereditary optic neuropathy reported from India is Leber's hereditary optic neuropathy (LHON), with a prevalence of approximately 1:50,000. Study from England puts the prevalence of disease at 3.22 in 1,00,000. They further report a prevalence of 11.82 in 1,00,000 individuals having a primary LHON related mitochondrial DNA (mtDNA) mutation (Chinnery et al., 2000). The pathogenetic mtDNA mutations for LHON are m.14484T>C (p.Met64Val) in MT-ND6 gene, m.11778G>A (p.Arg340His) in MT-ND4 gene, and m.3460G>A (p.Ala52Thr) in MT-ND1 gene, with locus 11,778 carrying the worst prognosis for vision, and locus 14,484 having the best prognosis. In a study from India, the frequency of m.11778G>A (p.Arg340His) was estimated at 29.22%, higher than reports of frequency of m.14484T>C (p.Met64Val) (4.2%) (Khan et al., 2013).
The type of mutation and its penetrance may vary among different haplogroups, with certain groups having protective or deleterious effects (Kumar et al., 2012; Khan et al., 2017). Apart from penetrance, the amount of phenotypic expression of disease also depends upon degree of heteroplasmy, which correlates with the risk of visual loss (Chinnery, Andrews, Turnbull, & Howell, 2001). Authors have demonstrated that sex bias and heteroplasmy play strong role in Indian cases of LHON, with a 20.3% incidence of heteroplasmy for m.11778G>A (p.Arg340His) mutation among families studied. These heteroplasmic families may show a protective effect on visual function (Khan et al., 2017).
Another aspect which affects the risk of vision loss in LHON patients is the associated haplotype in patients. European LHON patients with the primary mutations show increased risk in haplogroups J1, J2, and K, while haplotype H was found to have a protective role for m.11778G>A mutation (Hudson et al., 2007). However, one LHON study from India has shown that there is no such similar association between m.11778G>A (p.Arg340His) mutation and mtDNA haplogroups. Another study in Indian patients carrying the m.14484T>C (p.Met64Val) mutation found associations with haplogroups F1c1, M31a, U2a, etc., in contrast to European data of association with the J haplogroup (Thangaraj et al., 2006; Howell et al., 2003).
Moreover, distinct sequence variations in Indian pedigrees suggest that the primary mutations might have arisen independently in different mitochondrial haplogroup backgrounds, without any specific association with any haplogroup (Khan et al., 2017; Thangaraj et al., 2006). This again can be attributed to consanguinity, and strict endogamy marriage practices (Thangaraj et al., 2006).
Apart from the primary mutations commonly resulting in LHON (Jiang et al., 2016), several secondary mitochondrial mutations known as secondary mutations may also affect pathogenesis, as has been demonstrated in Indian studies as well (Kumar, Tanwar, Saxena, Sharma, & Dada, 2010; Kumar et al., 2012). Novel mutations have been described by Kumar et al. in MT-CO3, MT-CYB, and MT-ATP6 genes (Kumar et al., 2012). MT-CYB gene mutations may affect the transmembrane region of the cytochrome b protein and lead to defective interaction with other proteins. MT-ATP6 gene mutations may cause dysfunction of the ATP synthase.
5 CONGENITAL OCULAR MOTILITY DISORDERS
5.1 BPES syndrome
Autosomal dominant mutation in FOXL2 gene on chromosome 3q has been found to be associated with a syndrome characterized by blepharophimosis, ptosis, telecanthus, and lid phimosis has been reported (Kumar, Babu, Raghunath, & Venkatesh, 2004). A novel polyalanine expansion in FOXL2 has been reported from India, which leads to associated ovarian dysfunction in BPES patients (Nallathambi et al., 2007).
5.2 Congenital cranial dysinnervation disorders (CCDDs)
These constitute a group of congenital ocular muscle disorders, with a neurological basis. Among them, congenital fibrosis of extraocular muscles (CFEOM) is a dysinnervation strabismus syndrome presenting with variable angles of strabismus, ptosis and pupilloplegia. CFEOM is caused by pathogenic variants in the genes encoding KIF21A, PHOX2A, TUBB3, and TUBB2B. The selective absence of global layers of all extraocular muscles has been reported from North India, suggesting separate developmental pathways for different types of fibers of the extraocular muscles (Saini, Sharma, Gaur, & Singh, 2017).
Complete progressive external ophthalmoplegia (CPEO) or Kearns-Sayre syndrome, are caused due to mitochondrial DNA mutations. In a study from South India, complete mitochondrial genome analysis showed mutations in genes of tRNA, rRNA, NADH dehydrogenase, cytochrome B and cytochrome oxidase I, II, and III. Of the biopsies which did not show the pathognomonic ragged-red muscle-fiber morphology, 90% still had mitochondrial gene mutations (Sundaram et al., 2011).
Indian scientists have developed a cybrid containing the mitochondrial DNA deletion from a patient with Kearns-Sayre syndrome. The treated cells showed translation of the polycistronic RNA1-encoded mRNAs in mitochondria and the increase in respiration function of the cells by 90% of normal (Mahato, Jash, & Adhya, 2011). The same group of authors have also demonstrated ATP-dependent import of human cytoplasmic tRNA into human mitochondria in vitro, which was found to support mitochondrial protein synthesis and may potentially be of therapeutic application (Mahata, Bhattacharyya, Mukherjee, & Adhya, 2005).
5.3 Disorders associated with nystagmus
5.3.1 Congenital nystagmus
Pedigree analysis studies have revealed different modes of inheritance for congenital nystagmus (CN), of which the X-linked form of CN (XLCN) is the most prevalent (Self & Lotery, 2006). Radhakrishna et al. first confirmed FRMD7 gene mutations in XLCN cases in India and also identified novel missense mutation, c.A917>G (p.Gln305Arg) in a multigenerational family, which leads to an energetically unstable protein (Radhakrishna et al., 2012). Another study by Gupta et al. revealed a novel c.556A>G (p.Met186Val) mutation in FRMD7 gene exon causing XLCN. They also studied the protein homology modeling, and inferred that this mutation may lead to change in protein conformation and loss of interactions between different protein domains (Gupta et al., 2015).
5.3.2 Systemic disorders showing nystagmus
A common form of ataxia which may be associated with nystagmus is spinocerebellar ataxias (SCAs). In India, SCA type 2 and type 1 are the most prevalent (Sinha et al., 2004; Krishna et al., 2007). Authors from the country have demonstrated that optokinetic nystagmus present in SCA patients may be used as a very sensitive bedside clinical tool to predict early symptomatic or presymptomatic SCA1, SCA2, and SCA3 in genetically related siblings.
6 GENETICS OF CONGENITAL AND JUVENILE GLAUCOMA
CYP1B1 gene mutations have been studied extensively in glaucoma. Three novel mutations in CYP1B1 c.71T>G (p.Leu24Arg), g.4373T>C (p.Phe190Leu), and p.Gly329Asp, have been reported from Northern India (Tanwar et al., 2009, p. 1). Sturge-Weber syndrome (SWS) associated with CYP1B1 was reported by the same group (Tanwar et al., 2010). Further, Indian scientists have reported that the pathologic state of the carrier of CYP1B1 gene mutation is affected by the allelic state of the gene (Banerjee, Chakraborty, Chakraborty, Chakrabarti, & Ray, 2016). They further correlated CYP1B1 and Myocilin gene, MYOC, and found that mutant CYP1B1 lacking the 17β estradiol activity can cause MYOC upregulation, affecting glaucoma pathogenesis (Mookherjee, Acharya, Banerjee, Bhattacharjee, & Ray, 2012). Myocilin gene, MYOC, mutations have largely been implicated in juvenile open angle glaucoma (JOAG). Indian data has shown a higher prevalence of MYOC mutations in familial cases of JOAG. The most common variant reported being c.1099G>A (p.Gly367Arg) (Gupta et al., 2018).
7 DATABASES AND CONSORTIA
Genomic sequencing consortia funded by the government such as GUaRDIAN, (GUaRDIAN Consortium, Sivasubbu, & Scaria, 2019), Indian Genetic Disease Database (IGDD) (Pradhan et al., 2011) are helping us understand the population genetics. Specifically, IGDD website http://www.igdd.iicb.res.in/home.htm enlists the specific mutations found in the Indian population. THE GUaRDIAN program provides the bridge connecting clinicians and scientists, allowing patients with rare diseases access to genomic sequencing in a government funded program (http://guardian.meragenome.com/home). Similarly, the INDIGEN program provides access to the sequenced genomes from across India (http://clingen.igib.res.in/indigen/index.php). Among the initial studies, Indian Genomic Variation Consortium, reported the SNPs associated with CYP1B1 gene, important in glaucoma, and showed its association with ethnic diversity distribution in the country (Indian Genome Variation Consortium, 2008). India Allele Finder, curated and presented data from the 1,000 genome browser (Zhang, James, Shukla, Girisha, & Paciorkowski, 2017). Among the multiple forums for rare diseases in India, organizations such as the Organization for Rare Diseases in India, ORDI, have developed a platform for all information regarding rare diseases in India, catering to the patients. It helps maintain a registry for patients with rare diseases. Forums like these form a connection between the government, patients and healthcare infrastructure overall (Rajasimha et al., 2014). The Asia Eye Genetics Consortium, with its India chapter aims to address the eye genetics needs in India (Iwata, 2016).
8 CHALLENGES IN PRACTICE
There are a significant number of individuals affected by genetic eye diseases in India as discussed in the sections above. However, when compared to the overall health priorities of the country, it features low in the list given the higher prevalence of other communicable and noncommunicable diseases. Infrastructure, social practices, and the gene pool all add to the challenges faced in practice.
8.1 Health economics
Genetic eye diseases often result in severe progressive vision loss. The associated visual impairment, leads to higher Disability-Adjusted Life Years (DALY) (Menon et al., 2019).
Further, the emphasis on expenditure for rare diseases is not a thrust area in India. However, this economic challenge aside, the Indian government funding toward scientific research, and basic sciences projects has been growing by 8.8% over the years (Dandona et al., 2017).
8.2 Awareness, social beliefs, and consanguinity
Social beliefs and consanguinity are among the heart of the challenges faced. Lack of awareness among the population (Sen et al., 2008) that an inherited condition can be the cause of visual impairment is an important factor. While marriages within a community are a commonplace, availability of information, and effective genetic counseling can help put things in perspective for the affected group (Clarke, Combs, Black, & Hall, 2015).
8.3 Diagnostic facilities
Cost and access to genomic diagnostics remains a challenge. While gaining popularity and momentum, knowledge of genomic sequencing among health care providers is a challenge as well. However, with advances in biotechnology and potential lowering of costs, it is hopefully likely to change for the better in the future.
8.4 Healthcare infrastructure
In a busy eye clinic, which most often is the case, and without therapeutic options available, patients with genetic eye disease typically may not receive adequate attention. Addressing all concerns, from explaining the nature of diseases, prognosis, rehabilitation options available requires time, which may not be possible most often with lack of dedicated personnel.
Even if available, in the absence of therapy, and formidable cost of diagnostic sequencing, many patients decline from getting them done. Visual rehabilitative services, low visual aids which are potentially most important for the care of such patients, are typically scarce.
9 SOLUTIONS TO CHALLENGES ENCOUNTERED
9.1 Dedicated centers of excellence for ophthalmic genetics
Centers of excellence for eye genetics, with a strong referral system are needed. At such centers, availability of NGS, genetic counselors, visual rehabilitation, and facilities for translational eye research should be made available. This would include training of eye specialists, genetic counselors, and low vision specialists in this area of ophthalmology. Setting up translational research facilities, with emphasis on clinician–scientist collaborative work will further aid development of solutions tailored to the indigenous population, most relevant in this era of precision medicine.
9.2 Telemedicine and telegenetics
Counseling can be done most effectively with dedicated time, in the absence of personnel at a certain location, teleophthalmology, and telegenetics may provide a plausible solution (Zierhut, MacFarlane, Ahmed, & Davies, 2018). Telecommunication, access to smartphones and internet has reached the deepest interiors of India. Telemedicine and telegenetics, therefore, holds immense potential. Patient support groups, although limited at present, can surely help disseminate information, as well as provide emotional support.
9.3 Eye research funding opportunities in India
Indo-US Collaborative initiative, led by the Department of Biotechnology (DBT, India) and National Eye Institute, United States—is an important grant toward eye related collaborative research. Further, the India Alliance, Indian Council of Medical Research (ICMR), Council for Scientific and Industrial Research (CSIR), and other scientific bodies offer funding opportunities.
10 ADVANCES IN BASIC SCIENCES EYE RESEARCH IN INDIA
In the era of precision medicine, the solutions for genetic disease need to be tailored to the Indian population. The scientific milieu in India is advancing swiftly. Funding programs, government schemes, as well as enhanced knowledge sharing among scientists are depictive of a bright future for ophthalmic genetics, both literally and figuratively. In this section we will review the recent advances in basic sciences eye research in India.
10.1 Stem cells
Inducible pluripotent stem cells (iPSC) based eye research is a growing area in India. Both corneal and retinal organoids have been studied by scientists in the country. An efficient method for development of mini-corneal organoids, derived from iPSC, in three-dimensional, suspension medium has been reported from South India (Susaimanickam et al., 2017). This was in contrast to two-dimensional and fluorescence activated cell sorting (FACS) methods described previously. This has opened new avenues of corneal research. It may serve as an important corneal disease model, as well as be used to treat ocular surface diseases such as limbal stem cell deficiency (LSCD). Efficient method of development of iPSC-derived retinal pigment epithelial (RPE) cells has been developed and reported from the country (Surendran, Rathod, & Pal, 2019). Indian groups have developed and reported generation and characterization of iPSC from patients with different retinal dystrophies (Konala, Nandakumar, Battu, & Pal, 2020). These reports show the growing regenerative research milieu.
10.2 Gene therapy
The ICMR, has laid out guidelines for Gene Therapy Research in India recently. Indian groups in collaboration are exploring gene therapy using adeno-associated virus (AAV) vectors as a vector to treat corneal inflammation (Gupta et al., 2018). Genomic editing using CRISPR/Cas9 although explored in nonophthalmic conditions by Indian scientists, is likely to see more applications in ophthalmic genetics in near future (Acharya et al., 2019).
11 CONCLUSION
In conclusion, ophthalmic genetics in India has found footing in the past decades, and the decade to come is likely to see remarkable growth. Awareness of NGS, both among ophthalmologists and patients, as well as spreading knowledge of hereditary nature of diseases among the population are likely to play an important role. Genetic mutation analysis from databases have revealed a distinct genotypic variation in our country and the role of precision medicine is being increasingly realized. Advances in cutting edge genomic editing research, stem cell research, and supportive government funding are promising pillars for developing therapies for patient benefit.
LITERATURE SEARCH
Literature search was done using Pubmed Medline, Cochrane library Database, and Scopus using the terms eye genetics, corneal dystrophies, retinal dystrophies, hereditary optic neuropathies, congenital cranial dysinnervation disorders; and selecting location ID as India where applicable. All relevant articles were included with priority given to randomized control trials and prospective studies. Reference lists from selected articles were further reviewed to get articles not included in the electronic database.