Genetic overlap between epilepsy and schizophrenia: Evidence from cross phenotype analysis in Hong Kong Chinese population
Abstract
Epilepsy and schizophrenia are common and typical neurological or mental illness respectively, and sometimes they comorbid in the same patients, however the underlying genetic relationship between the two brain diseases is still not fully understood. To investigate the possible genetic contribution to their comorbidity, we performed polygenic risk score (PRS) analyses and genetic correlation estimation so as to identify the overall genetic overlap between the two diseases. The global schizophrenia PRS is strongly associated with schizophrenia phenotype in Hong Kong population (odds ratio = 1.7, p = 2.26E-16), and focal epilepsy PRS is moderately associated with epilepsy phenotype in Hong Kong population (odds ratio = 1.14, p = 0.013). However the disease-specific PRS can only predict its own well-matched phenotype but not the other ones (p > 0.05). This pattern is further supported by non-significant pairwise genetic correlation and insufficient statistical power for PRS association from the cross-phenotype analyses. Our study reveals there's limited shared genetic aetiology between schizophrenia and epilepsy, and thus supports a model of shared environmental factors to explain the comorbidity between the two phenotypes.
1 INTRODUCTION
Epilepsy and schizophrenia are two common brain-involved disorders. Patients with epilepsy are at increased risk of developing psychosis compared with the general population (Chen et al., 2016; Tellez-Zenteno, Patten, Jette, Williams, & Wiebe, 2007; Torta & Keller, 1999; Vuilleumier & Jallon, 1998) and some forms of seizures may mimic the symptoms of psychosis (Sethi & Emery, 2013; Tucker, 1998); therefore better understanding of epilepsy might also provide insights on the understanding of mental illness like schizophrenia, and vice versa. One example is from brain image studies, which revealed hippocampus volume is probably related to both epilepsy and schizophrenia (Jack, 1994; Nelson, Saykin, Flashman, & Riordan, 1997). Another example comes from a family-based study that gave evidence of two-fold increase in the risk of developing psychosis for individuals with a parental history of epilepsy (Clarke et al., 2012). Moreover, comorbidity analysis from previous epidemiology study in Chinese (Chang et al., 2011) found higher incidence of epilepsy in schizophrenia cohort than non-schizophrenia cohort (6.99 vs. 1.19 per 1,000 person-years) and also higher incidence of schizophrenia in epilepsy cohort than non-epilepsy cohort (3.53 vs. 0.46 per 1,000 person-years), hence showed they may relate to each other at disease susceptibility.
However, from the perspective of genetics, few susceptibility loci have been found to predispose to both two diseases (Welter et al., 2014). Moreover, genetic overlaps between them are not clear due to different hypothesis in previous candidate-gene based research. Findings from previous genome-wide association studies (GWAS) for epilepsy and schizophrenia that are stored in GWAS Catalog (https://www.ebi.ac.uk/gwas/home) share very few hits for both. This can be ascribed to limited power of univariate SNP analysis, given GWAS variants are mostly with small effect sizes on the phenotypes (Park et al., 2010). Nevertheless, GWAS provide good resources for secondary analyses that aim to leverage the power of multiple phenotypes, or establish the genetic relationship between them (Bulik-Sullivan, Finucane, et al., 2015; Solovieff, Cotsapas, Lee, Purcell, & Smoller, 2013). Among different data mining approaches, PRS analysis is widely used to search for shared genetic aetiology between two datasets on the same phenotype or different phenotypes (Purcell et al., 2009; Chatterjee, Shi, & Garcia-Closas, 2016). Regarding to epilepsy and schizophrenia, large-scale GWAS meta-analyses have already been reported for them respectively (International League Against Epilepsy Consortium on Complex Epilepsies. Electronic address 2014; Ripke et al., 2014) and can provide necessary training dataset for PRS construction. In addition, our previous GWAS reports on both schizophrenia and epilepsy can serve as independent target dataset for testing PRS association (Guo et al., 2012; Wong et al., 2014).
In this study, we systematically estimated the overall genetic sharing by using publicly available summary statistics and in-house genome-wide raw genotypes for both epilepsy and schizophrenia. We focused mainly on PRS analysis in our Chinese population, together with genetic correlation analysis between different datasets, so as to establish the genotype-phenotype relationship across these two diseases.
2 METHODS
2.1 Data collection and preprocessing
For training datasets, we collected summary statistics (SNP ID, alleles, p-values, odds ratio (OR), and standard errors) from GWAS meta-analyses previously reported by International League Against Epilepsy (ILAE) consortium on complex epilepsies (International League Against Epilepsy Consortium on Complex Epilepsies. Electronic address 2014) and Psychiatric genomics consortium (PGC) (Ripke et al., 2014), respectively. The latest meta-analysis result on schizophrenia for 49 Caucasian population cohorts (in total 33,640 cases and 43,456 controls) was downloaded from PGC public repository (http://www.med.unc.edu/pgc/). Regarding to epilepsy, we re-performed fixed-effect meta-analyses for all epilepsies (including both genetic generalized epilepsy (GGE) and focal epilepsy) using Caucasian samples (7,019 cases and 19,935 controls) and for focal epilepsy separately (4,050 cases and 19,935 controls), since Hong Kong cohort was comprised of focal epilepsy patients only.
For target datasets, we retrieved raw genotype data from two previous GWAS done in our team, for epilepsy or schizophrenia in Hong Kong (HK) Chinese population, respectively (Guo et al., 2012; Wong et al., 2014). Samples from both GWAS were genotyped on the Human610-Quad BeadChip (Illumina, Inc., San Diego, CA). Both datasets have been imputed to 1,000 Genome reference panel using the same protocol. Moreover, to eliminate the influence of different controls on polygenic score estimation for two target datasets, we kept only 1661 shared controls between them. For HK GWAS datasets after standard quality control, we estimated the population structure by principal component (PC) analysis using linkage disequilibrium (LD) pruned SNPs (Patterson, Price, & Reich, 2006); then we generated univariate SNP-level summary statistics by running PLINK logistic regression between binary phenotype and additive minor allele copies (0, 1 or 2) (Purcell et al., 2007), while adjusting for significant PCs determined by R package “EigenCorr” (Lee, Wright, & Zou, 2011). To reduce mismatched markers between datasets from different resources, we restricted genetic markers to HapMap3 variants only. A python script in LD Score Regression (LDSC) package (https://github.com/bulik/ldsc) was used to filter out unqualified variants from either training or target datasets. Table 1 lists the final data we used in this study.
2.2 PRS association analysis
For each of two target datasets from Hong Kong epilepsy GWAS (HKEPI) and Hong Kong schizophrenia GWAS (HKSCZ), we calculated its polygenic risk scores using all epilepsies meta-GWAS (EPIall), focal epilepsy meta-GWAS (EPIfocal) or schizophrenia meta-GWAS (SCZ) as training dataset (Table 1). In brief, independent SNPs were generated by LD clumping (r2 < 0.1) in target dataset and then summed up after weighting each by its effect size in training dataset. A logistic regression was then used to test the association between each PRS and the corresponding phenotype (binary status of disease), with adjustment of the same principal components from univariate SNP analysis. To select different groups of SNPs in PRS, we switched inclusion p-value threshold from 0 to 0.5 by 0.005 increment and chose the best-fit model according to its variance explained (Nagelkerke's pseudo R2). PRSice was used to perform the calculation, model fitting and visualization (Euesden, Lewis, & O'Reilly, 2015). In order to present a more meaningful effect size, the raw PRSs were standardized to have distribution with mean 0 and variance 1. ORs relative to one standard deviation increase were computed using beta estimates from logistic regression model newly fitted for standardized PRS. This transformation and modeling was done manually in R program (https://www.r-project.org/).
| Dataset | Disease prevelance | Resources | Data after processinga |
|---|---|---|---|
| EPIall | 0.005 | Summary statistics from LAE meta-analysisb | 7019 cases, 19935 controls; ∼1.1M SNPs |
| EPIfocal | 0.003 | Summary statistics from ILAE meta-analysis | 4050 case, 17481 controls; ∼1.1M SNPs |
| HKEPI | 0.003 | Raw genotypes from HK GWAS | 522 cases, 1661 controls; ∼800 K SNPs |
| SCZ | 0.01 | Summary statistics from PGC meta-analysisc | 33356 cases, 43724 controls; ∼1.1M SNPs |
| HKSCZ | 0.01 | Raw genotypes from HK GWAS | 377 cases, 1661 controls; ∼800 K SNPs |
- a Both sample filtering and variant filtering were adopted (see method part for detail).
- b Caucasian population cohorts were included for re-running meta-analysis.
- c Meta-analysis using all 49 Caucasian cohorts. ILAE, international league against epilepsy; PGC, psychiatric genomic consortium.
2.3 Heritability and genetic correlation
LDSC approach (Bulik-Sullivan, Finucane, et al., 2015) was used to calculate heritability at liability scale for each dataset (SCZ, EPIall, EPIfocal, HKSCZ, and HKEPI), also genetic co-variance and correlation between each pair of the five datasets. Latest python script ldsc.py (https://github.com/bulik/ldsc, May 2017) was applied for all calculations, with HapMap Caucasian or East Asian reference panel to adjust for LD pattern between genetic markers.
2.4 Power estimation
We used “polygenescore” function to calculate the power of PRS association analyses conducted in this study (Dudbridge, 2013), with R code downloaded from https://sites.google.com/site/fdudbridge/software/. For each pair involving one training dataset and one target dataset, disease prevalences (K1, K2), sample size (n1, n2) and sampling ratios (P1, P2) were from Table 1, while heritability in training dataset (σ12) and genetic co-variance between two datasets (σ12) were acquired by calculation in above section. Numbers of independent markers (m) were estimated by LD clumping at r2<0.1 for GWAS genotypes from target dataset, and proportion of markers with no effect (π0) was defined as percentage of independent SNPs with p-value >0.05 in summary statistics from training dataset. Type I error rate (alpha) was set at 0.001.
3 RESULTS
Genomic inflation factors (λ) in training datasets are 1.61 for SCZ, 0.99 for EPIall, and 0.92 for EPIfocal; in target dataset, λ is 1.02 and 1.04 for HKSCZ and HKEPI respectively after adjusting for two significant PCs. The bar charts (Figure 1) show the model fitting and associations of PRS from various p-value thresholds on the different datasets. First, schizophrenia PRSs are strongly associated with the status of schizophrenia in HK population (Figure 1a), with variance explained (pseudo R2) at 5.1%, and p-value at 2.26E−16 for the best-fit model; however, SCZ PRS does not predict HK epilepsy status at all (Figure 1b) since all p-values are above 0.05. Second, all epilepsies PRSs are not associated with either HKSCZ (Figure 1c) or HKEPI (Figure 1d). Third, though focal epilepsy PRSs do not predict the status of HKSCZ either (Figure 1e), the best-fit PRS is moderately associated with HKEPI (variance explained = 0.4%, p = 0.013) (Figure 1f). Furthermore, stratified analysis on HKEPI reveals EPIfocal PRS associates mostly with HK focal epilepsy patients documented hippocampal sclerosis lesion (variance explained = 1.9%, p = 0.001), but not with other sub-phenotypes (Figure S1).
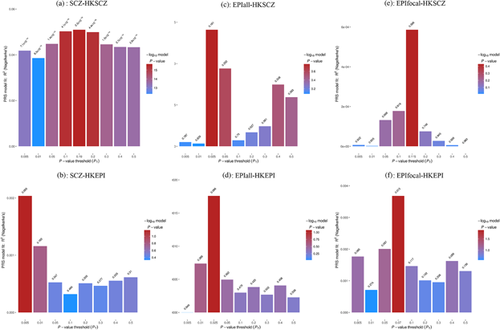
With selection of three fixed thresholds (0.005, 0.05, and 0.5) and the one from best-fit model, odds ratios computed using standardized PRS are shown in Table 2. SCZ PRS consistently increases the risk of schizophrenia in Hong Kong population at all different thresholds (OR > 1.6, p < 1.0E-13), and focal epilepsy PRS only slightly increases the risk of epilepsy in Hong Kong (OR = 1.14, p = 0.013). No pairs crossing two different diseases are found with statistical evidence of PRS association, indicating there is limited genetic overlapping between epilepsy and schizophrenia.
| SCZ PRS | EPIall PRS | EPIfocal PRS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Dataset | Threshold | #SNPs | OR (99.9%CI) | p | #SNPs | OR (99.9%CI) | p | #SNPs | OR (99.9%CI) | p |
| HK SCZ | 0.5 | 47610 | 1.62 (1.31–1.99) | 2.77E-14 | 48486 | 1.06 (0.87–1.29) | 0.363 | 44756 | 1.00 (0.82–1.22) | 0.983 |
| 0.05 | 15702 | 1.65 (1.33–2.04) | 1.38E-14 | 11977 | 1.07 (0.88–1.31 | 0.252 | 10701 | 1.03 (0.84–1.26) | 0.668 | |
| 0.005 | 4938 | 1.61 (1.30–1.98) | 7.13E-14 | 1819 | 0.98 (0.81–1.20) | 0.787 | 1538 | 0.99 (0.81–1.21) | 0.922 | |
| Best fit | 30796 | 1.70 (1.37–2.10) | 2.26E-16 | 6973 | 1.09 (0.89–1.33) | 0.161 | 19544 | 0.95 (0.77–1.16) | 0.366 | |
| HK EPI | 0.5 | 47701 | 1.06 (0.88–1.26) | 0.310 | 48486 | 1.03 (0.86–1.24) | 0.536 | 44761 | 1.08 (0.91–1.30) | 0.139 |
| 0.05 | 15718 | 1.05 (0.88–1.26) | 0.347 | 11976 | 1.05 (0.88–1.26) | 0.362 | 10704 | 1.10 (0.92–1.32) | 0.067 | |
| 0.005 | 4941 | 1.10 (0.93–1.32) | 0.065 | 1819 | 1.00 (0.83–1.19) | 0.949 | 1536 | 1.10 (0.92–1.32) | 0.085 | |
| Best fit | 4941 | 1.10 (0.93–1.32) | 0.065 | 6973 | 1.10 (0.92–1.31) | 0.086 | 13801 | 1.14 (0.96–1.37) | 0.013 | |
- Odds ratio (OR), 99.9% confidence interval (99.9%CI) and p-values were calculated for standardized PRS by logistic regression with adjustment for two PCs. #SNPs, number of SNPs included in PRS construction given a significance threshold.
Table 3 gives the estimated heritability for each dataset and genetic correlation for each pair of datasets, using summary statistics from common SNPs. Only two pairs are found with significant genetic correlations (SCZ and HKSCZ, R2 = 0.39, p = 4.9E-10; EPIall and EPIfocal, R2 = 0.91, p = 3.7E-275). Similar to PRS association, we do not find any evidence of shared genetic aetiology between schizophrenia and epilepsy, even with up-to-date largest meta-analysis datasets (SCZ and EPIall, R2 = 0.08, p = 0.164).
| Dataset1 | Dataset2 | σ12 | σ22 | σ12 | R2 | p |
|---|---|---|---|---|---|---|
| SCZ | HKSCZ | 0.25 (0.01) | 0.23 (0.24) | 0.23 (0.02) | 0.39 (0.06) | 4.90E-10 |
| SCZ | HKEPI | 0.25 (0.01) | 0.24 (0.13) | 0.04 (0.02) | 0.06 (0.04) | 0.116 |
| EPIall | HKSCZ | 0.08 (0.01) | 0.23 (0.24) | −0.01 (0.05) | −0.06 (0.38) | 0.868 |
| EPIall | HKEPI | 0.08 (0.01) | 0.24 (0.13) | 0.03 (0.04) | 0.13 (0.23) | 0.575 |
| EPIfocal | HKSCZ | 0.12 (0.01) | 0.23 (0.24) | −0.05 (0.05) | −0.42 (0.76) | 0.58 |
| EPIfocal | HKEPI | 0.12 (0.01) | 0.24 (0.13) | −0.01 (0.05) | −0.06 (0.27) | 0.818 |
| HKSCZ | HKEPI | 0.23 (0.24) | 0.24 (0.13) | 0.08 (0.16) | 0.20 (0.34) | 0.553 |
| SCZ | EPIall | 0.25 (0.01) | 0.08 (0.01) | 0.02 (0.01) | 0.08 (0.06) | 0.164 |
| SCZ | EPIfocal | 0.25 (0.01) | 0.12 (0.01) | 0.01 (0.01) | 0.04 (0.05) | 0.363 |
| EPIall | EPIfocal | 0.08 (0.01) | 0.12 (0.01) | 0.14 (0.02) | 0.91 (0.03) | 3.70E-275 |
- σ12 and σ22 are heritability (in liability scale) for dataset1 and dataset2; σ12 and R2 are genetic covariance and genetic correlation between dataset1 and dataset2. Numbers in bracket are standard errors for each estimate.
Finally, we plugged epidemiological parameters in Table 1 and LDSC calculated genetic parameters in Table 3 into the function of power calculation for PRS association in Table 2. As expected, current training dataset from schizophrenia is well powered for predicting HK SCZ at all different thresholds (Table 4). The comparatively lower power (<0.5) of predicting HK EPI by SCZ PRS reflects limited genetic correlation between epilepsy and schizophrenia. All other power estimates are under 0.1 (Table 4). This may be due to much smaller sample size and SNP-based heritability from training dataset (EPIall or EPIfocal), or inaccurate genetic covariance (with wide standard error) between two datasets with relatively low sample size in either one or both.
| Threshold | |||||
|---|---|---|---|---|---|
| Phenotype pairs | Parameters1 | 0.005 | 0.05 | 0.5 | Best fit |
| SCZ, HKSCZ | 0.01, 0.01, 77080, 2038, 0.43, 0.18, 0.25, 0.23, 56162, 0.88 | 1.000 | 1.000 | 1.000 | 1.000 |
| SCZ, HKEPI | 0.01, 0.003, 77080, 2138, 0.43, 0.24, 0.25, 0.04, 59821, 0.75 | 0.284 | 0.405 | 0.364 | 0.284 |
| EPIall, HKSCZ | 0.005, 0.01, 26954, 2038, 0.26, 0.18, 0.08, −0.01, 60471, 0.8 | 0.001 | 0.001 | 0.002 | 0.001 |
| EPIall, HKEPI | 0.005, 0.003, 26954, 2183, 0.26, 0.24, 0.08, 0.03, 55595, 0.95 | 0.072 | 0.066 | 0.045 | 0.071 |
| EPIfocal, HKSCZ | 0.003, 0.01, 23985, 2038, 0.17, 0.18, 0.12, −0.05, 53940, 0.86 | 0.011 | 0.032 | 0.058 | 0.042 |
| EPIfocal, HKEPI | 0.003, 0.003, 23985, 2183, 0.17, 0.24, 0.12, −0.01, 50375, 0.95 | 0.004 | 0.003 | 0.003 | 0.003 |
- Parameters in order for K1, K2, n1, n2, P1, P2, σ12, σ12, m, and π0 respectively.
4 DISCUSSION
Epilepsy and schizophrenia are two common diseases belonging to the same broad category of brain disorders, which cause huge economic burden for the patient, his/her families and the society. Our study reveals the potential of using PRS to predict the same phenotype (especially on schizophrenia); in addition it suggests no significant amount of genetic overlap may exist between epilepsy and schizophrenia.
Secondary analyses on GWAS dataset are still providing new insights for complex phenotypes (Barabasi, Gulbahce, & Loscalzo, 2011; Bulik-Sullivan, Finucane, et al., 2015; Chatterjee et al., 2016). The more advanced data mining approaches can help to reveal hidden genetic signals for one phenotype or multiple phenotypes together (Cantor, Lange, & Sinsheimer, 2010; Yang et al., 2012). We previously performed multilevel GWAS analysis to search for overlapping functional units (genic SNPs, genes, or gene-sets) across different complex diseases (Gui, Kwan, Sham, Cherny, & Li, 2017), but found no significant sharing between epilepsy and schizophrenia (data not shown). To further examine the genetic relationship between schizophrenia and epilepsy, we resorted to PRS analysis and genetic correlation analysis in this study. These two approaches have been independently established and applied in a few complex diseases or traits before (Bulik-Sullivan, Finucane, et al., 2015; Purcell et al., 2009; Chatterjee et al., 2016). In our study the results from them agree mostly with each other, as both support the close relationship between global schizophrenia and Hong Kong schizophrenia, but not across epilepsy and schizophrenia. Given the nominal effect of SCZ PRS on our epilepsy patients (OR>1, p < 0.1) and <80% statistical power across the two phenotypes, we cannot completely rule out the possibility of elevating risk of epilepsy through schizophrenia. Nevertheless, the lack of genetic overlap between these two disorders is further supported by a recent finding that reveals substantial genetic difference between neurological and psychiatric disorders (Anttila et al., 2016). The genetic correlation coefficients between different kinds of epilepsy and schizophrenia are all reported at near-zero level. Though an earlier study using data from PGC1 and ILAE meta-analyses identified significant correlation between all epilepsies and schizophrenia (Vonberg & Bigdeli, 2016), we did not verify it when excluding Hong Kong cohort (schizophrenia patients served as controls to epilepsy patients in GWAS) from ILAE meta-analysis.
In comparison to clear relationship within schizophrenia datasets or across schizophrenia and epilepsy datasets, the pattern within epilepsy datasets is more vague and arbitrary. From PRS analysis, we identified moderate association of EPIfocal risk (instead of EPIall risk) on Hong Kong focal epilepsy patients (especially for the sub-phenotype characterized by hippocampal sclerosis). However, there is no significant genetic correlation found between epilepsy training and target datasets. A few factors may explain the inefficiency or discrepancy observed for epilepsy. First of all, epilepsy training dataset has much lower sample size than schizophrenia, hence can identify limited polygenicity among common SNPs (Bulik-Sullivan, Loh, et al., 2015); accordingly, the explained heritability by the same SNP array is much lower than that for schizophrenia. Second, both epilepsy training dataset and target dataset have more heterogeneous phenotype than that for schizophrenia. Thirdly, the LD pattern in training dataset (Caucasian population) is different from targeting dataset (Asian population), hence may cause different extent of power reduction or bias estimate from either PRS prediction (Martin et al., 2017) or genetic correlation analysis (Bulik-Sullivan, Finucane, et al., 2015). All these factors contribute to the small power estimates (Table 4) for phenotype pairs involving epilepsy dataset and highlight the limitation of current study. Specifically, we have observed better model fitting (variance explained from 2.8% to 5.1%) for SCZ PRS on HKSCZ when switching from PGC1 to PGC2. Therefore improvement from these factors, either a magnitude increase of sample size, or homogenous phenotype selection, or well-matched population between training and target dataset, can be helpful to improve PRS association and risk prediction for epilepsy in later studies.
The lack of genetic sharing at genome-wide scale should redirect the effort to explain the comorbidity of epilepsy and schizophrenia to other potential mechanisms including environmental factors (Rudzinski & Meador, 2013). Neurological deficits at birth (including mental retardation and cerebral palsy) can increase the susceptibility to both diseases (Ottman, Annegers, Risch, Hauser, & Susser, 1996). Antecedent central nervous system injury (including stroke, trauma, and brain infection) may lead to hippocampal volume deficient and then cause the onset of epilepsy and schizophrenia (Schmitt, Malchow, Hasan, & Falkai, 2014). Other factors, like stress during development, depression, and anti-epileptic drug effects on mental state, are all important contributor to the comorbidity (Agrawal & Govender, 2011; Barnes & Paolicchi, 2008). Deeper understanding on the direct pathway from these environmental risk factors to two phenotypes has to await future studies.
ACKNOWLEDGMENTS
This study is supported by Small Project Funding from the University of Hong Kong (SPF 201409176131 to HS Gui). We would like to thank Dr. Timothy Mak for technical advice on running PRiSice, and Psychiatric Genomic Consortium for releasing their schizophrenia GWAS meta-analysis result available online.
CONFLICTS OF INTEREST
None declared.