Longitudinal heritability of childhood aggression
Abstract
The genetic and environmental contributions to the variation and longitudinal stability in childhood aggressive behavior were assessed in two large twin cohorts, the Netherlands Twin Register (NTR), and the Twins Early Development Study (TEDS; United Kingdom). In NTR, maternal ratings on aggression from the Child Behavior Checklist (CBCL) were available for 10,765 twin pairs at age 7, for 8,557 twin pairs at age 9/10, and for 7,176 twin pairs at age 12. In TEDS, parental ratings of conduct disorder from the Strength and Difficulty Questionnaire (SDQ) were available for 6,897 twin pairs at age 7, for 3,028 twin pairs at age 9 and for 5,716 twin pairs at age 12. In both studies, stability and heritability of aggressive behavioral problems was high. Heritability was on average somewhat, but significantly, lower in TEDS (around 60%) than in NTR (between 50% and 80%) and sex differences were slightly larger in the NTR sample. In both studies, the influence of shared environment was similar: in boys shared environment explained around 20% of the variation in aggression across all ages while in girls its influence was absent around age 7 and only came into play at later ages. Longitudinal genetic correlations were the main reason for stability of aggressive behavior. Individual differences in CBCL-Aggressive Behavior and SDQ-Conduct disorder throughout childhood are driven by a comparable but significantly different genetic architecture. © 2016 Wiley Periodicals, Inc.
INTRODUCTION
Aggression can be defined as a behavior intended to cause physical or emotional harm to others [Anderson and Bushman, 2002], but aggressive behaviors may also be beneficial to individuals by enabling them to survive through, for example, competition for limited resources [Lindenfors and Tullberg, 2011]. According to the Diagnostic and Statistical Manual of Mental Disorders, 5th Edition (DSM-5), childhood aggressive behavior is a criterion for disruptive behavior disorders such as oppositional defiant disorder (ODD) and conduct disorder (CD) [American Psychiatric Association, 2013]. Aggressive behavior is also implicated in neurodevelopmental disorders such as attention deficit hyperactivity disorder (ADHD) [Monuteaux et al., 2009; Hamshere et al., 2013] and antisocial personality disorder (ASPD) [Schaeffer et al., 2003; Nouvion et al., 2007]. Both low and high levels of aggression can be detrimental to survival and procreation, and it has been postulated that aggression is under stabilizing selection [Anholt and Mackay, 2012], implying that variation in aggression will show significant heritability. Heritability estimates for human aggressive behavior indeed tend to be high. In a meta-analysis by Miles and Carey [1997] of 24 different twin and adoption studies in children and adults, additive genetic effects were found to explain up to 48% of the variance. Similar estimates were observed in three related meta-analyses of anti-social behavior of children and young adults [Mason and Frick, 1994; Rhee and Waldman, 2002; Ferguson, 2010].
While genetic influences emerge in most behavioral genetic studies of aggression in humans, their magnitude varies across studies. Clearly, the heterogeneous nature of the aggression construct adds complexity as well as heterogeneity across age [Rhee and Waldman, 2002], and sex [Vierikko et al., 2003]. For age, the meta-analysis by Miles and Carey [1997] reported similar estimates for the contribution of environmental and genetic factors on aggression during childhood and adolescence. In contrast, two other meta-analyses by Ferguson [2010] and Rhee and Waldman [2002] demonstrated a decrease in standardized additive genetic effects on aggression between young children and adults.
Sex differences can also play a role. In general, boys are consistently rated as more aggressive than girls at all ages by their mothers, fathers, and teachers [Hudziak et al., 2003]. However, mean differences do not necessarily imply differing etiologies between sexes. Sex differences in genetic architecture may be quantitative (e.g., different heritabilities in boys and girls) or qualitative when different genes are expressed in the two sexes. When qualitative sex differences are of importance, one prediction is that the resemblance in siblings or twins of opposite sex is lower than expected based on the resemblance in same-sex siblings or twins. Vink et al. [2012] compared the correlations in same-sex and opposite-sex dizygotic twin pairs for aggression in 3–12-year-old twins (N > 30,000 children at 3 years, and N > 6,500 at age 12 years). There was no evidence for qualitative sex differences, as assessed by maternal ratings on the Child Behavior Check List (CBCL) aggression subscale, although sex differences in mean scores were large. Differences between raters may play a role in qualitative differences. Vierikki et al. [2003] found lower correlations in opposite-sex compared to same-sex DZ pairs for teacher ratings of aggression, thus suggesting sex-specific variation.
Some studies, but not all, find quantitative sex differences in the influence of genetic factors on aggression [Cadoret et al., 1995; Eley et al., 1999; Rhee and Waldman, 2002]. Eley et al. [1999] were unable to detect sex-differences in the genetic architecture for aggression assessed by the Strengths and Difficulties Questionnaires (SDQ) which was completed by parents in Swedish (aged 7–9) and British (aged 8–16) cohorts of 1,022 and 501 twin pairs, respectively. Other studies detected significant quantitative sex differences, but are inconclusive in regard to the direction of the effect [Silberg et al., 1994; Miles and Carey, 1997; van Beijsterveldt et al., 2003; Vierikko et al., 2003]. Silberg et al. [1994], for example, measured aggression by parental CBCL report in 1,264 twin pairs (aged 8–16 years) and found higher heritability estimates in boys, with a diminishing effect during adolescence. Vierikko et al. [2003] studied 1,651 Finnish twin pairs between the ages of 11 and 12 years, and observed lower heritability estimates in boys. The discrepancies in these findings could be due to large age ranges and differences in raters, as indicated by the study of Hudziak et al. [2003] who assessed aggression by multiple raters (mothers, fathers, and teachers) in over 6,000 Dutch twins aged of 3, 7, and 10 years old. Their study demonstrated sex differences in some, but not all raters and age groups. In addition, differences were diminished when the variance of aggressive behavior on which all raters agreed on was analyzed. These findings from individual studies echo the results of the meta-analysis by Miles and Carey [1997] which showed only slightly stronger effects of genetic influences in males than in age matched females.
Thus previous research has offered some indication for quantitative sex differences although of relative small effect, while other studies were unable to detect such differences. However, many studies were underpowered to detect small differences in heritability and the extent of quantitative sex differences and the dependency of such differences as a function of age remain unclear.
The stability of aggressive behavior in children generally is high. For example, in a 22-year longitudinal US-American study of over 600 subjects, their parents, and their children, aggression was found to be highly consistent from childhood until well into adulthood, and early aggressiveness was predictive of later serious antisocial behavior and self-reported physical aggression [Huesmann et al., 1984]. A number of behavior genetic studies have investigated the etiology of the stability in aggressive behavior. van Beijsterveldt et al. [2003] observed in a longitudinal Dutch sample of 3- to 12-year olds that stability across age intervals ranged from 0.41 to 0.77 and genetic factors accounted for most of this stability. A genetic longitudinal model suggested a dynamic developmental process consisting of transmission of existing genetic effects together with new genetic influences. The authors identified some modification of genetic influences by age and sex. At younger ages (3 and 7) heritability for aggression was around 60% and about the same in boys and girls, but at ages 10 and 12 years, heritability was 67% for boys, and 50–55% in girls. In contrast, for girls shared environmental factors were more important than for boys at 10 and 12 years (29%; whereas the estimates in boys were 16–19%). Similar results were found in two studies of overlapping samples of 10,038 British twin pairs between the ages 4 and 16 [Lewis and Plomin, 2015; Pingault et al., 2015]. In particular, Lewis and Plomin [2015] observed that genetic factors were the main source of stability in conduct problems between age 4 and 16, with constant effects of genetic influences throughout childhood (59% at age 4–61% at age 16). Similar, a study of twins aged 7–12-year-old found that stability of aggressive behavior was largely due to genetic factors and heritability was estimated between 76% and 84% [Haberstick et al., 2006]. Pingault et al. [2015] demonstrated the stable effect of genetic factors on conduct problems in a latent growth model and suggested that individual differences in the developmental trajectories might be under strong genetic influence. A further longitudinal study of 750 US-American twin pairs [Niv et al., 2013] during childhood (ages 9–10) and adolescence (ages 14–15) indicated that only part of the genetic factors that influence antisocial behavior (a latent factor that combined aggression and rule-breaking behavior) in adolescence overlaps with the genetic influences in childhood and that part of the genetic influence at ages 14–15 were adolescent specific. This study also demonstrated overlapping longitudinal genetic influence between aggression and rule-breaking behavior, indicating that a similar set of genes is influencing the developmental dynamics of both constructs. Thus, while some investigations have categorized subjects into different age groups [Niv et al., 2013], others have focused on developmental trajectory of the phenotype [Pingault et al., 2015] as well as on the influence of new and preexisting genetic factors [van Beijsterveldt et al., 2003].
Here we make use of the ongoing longitudinal data collections in the Twins Early Development Study (TEDS; Haworth et al. [2012]) and the Netherlands Twin Register (NTR; van Beijsterveldt et al. [2013]) to analyze data on aggressive behavior in children aged 7–12 years. The two cohorts assessed parental longitudinal ratings on aggression or conduct problems in very large samples, which enables the investigation of differences between sexes on the age-specific effects of genetic and environmental factors and the estimation of longitudinal genetic and environmental correlations. Importantly, sample sizes are sufficiently large to assess effects of common environment, shared by children growing up in the same family/household [Martin et al., 1978; Posthuma and Boomsma, 2000].
Aggression was assessed by two commonly used parent-rated instruments for children, the Child Behavior Check List- aggressive problems syndrome scale (CBCL; Achenbach and Rescorla [2010]) and the Strengths and Difficulties Questionnaires conduct problems scale (SDQ; Goodman [2001]) for NTR and TEDS, respectively.
These two datasets have been used in previous studies, most prominently Lewis and Plomin [2015], Pingault et al. [2015], and van Beijsterveldt et al. [2003]. However, previous studies on TEDS [Lewis and Plomin, 2015; Pingault et al., 2015] did not consider sex differences and the currently analyzed NTR dataset is now considerable larger than in the previous report [van Beijsterveldt et al., 2003] and thus has higher statistical power to detect differences across age and sex. Further, this is the first study which investigates these two large-scale longitudinal twin cohorts in a coherent framework. This study will also aid future genome wide association studies (GWAS) by investigating qualitative and quantitative sex differences as well as genetic stability over time, thus informing to what extent future GWA studies can collapse multiple studies across ages, sex, and instruments.
MATERIALS AND METHODS
Data
The Netherlands twin register (NTR)
The NTR was established in 1987 and collects data in twins and multiples from birth onwards [van Beijsterveldt et al., 2013]. Nationwide data collection is by mailed and/or online surveys. Parents of twins receive questionnaires when their twins are aged 1, 2, 3, 5, 7, 9/10, and 12 years of age. For the current analysis data of maternal rating at ages 7, 10, and 12 were analyzed for twin born between 1986 and 2005. To assess aggressive behavior the Child Behavior Checklist versions 4–18 and 6–18 was used [Achenbach and Rescorla, 2010]. The CBCL/6–18 is a revision of the CBCL/4–18. Because data were collected over the past 25 years both versions of the CBCL were used sequentially. Mothers were asked to rate the behavior of the child in the preceding 6 months on a 3-point scale; 0 if the problem item was not true of the child, 1 if the item was somewhat or sometimes true, and 2 if it was very true or often true. The syndrome scale Aggressive Behavior (AGG) was composed by adding the scores on syndrome-specific questions according to the 1991 profile [Achenbach and Rescorla, 2010]. AGG consists of 18 items. Data from subjects with more than three missing items were not included in the analyses. This occurred in less than 2.5% of the received questionnaires. Maternal ratings on AGG were available for 10,765 twin pairs at age 7, 8,557 twin pairs at age 9/10, and 7,176 twins pairs at age 12.
Twin early development study (TEDS)
TEDS was established in 1995 with three birth cohorts (1994–96) obtained from UK birth records. In infancy and early childhood, questionnaires were posted to parents and teachers (with permission from parents), and school achievement records were also obtained [Haworth et al., 2012]. Data were gathered from telephone and in person interviews as well as increasingly from online internet assessments. The measure used consistently at all ages and all sources (including the twins themselves beginning at age 10) is the Strength and Difficulties Questionnaire (SDQ; Goodman [1997, 2001]). For the current study parental (maternal or paternal) ratings were used. The SDQ is a 25-item questionnaire designed to measure common mental health problems during childhood and adolescence. Ratings are on a three-point scale. The conduct problem scale with five separate items was used to measure aggression within TEDS. Parental ratings were available for 6,897 twin pairs at age 7, 3,028 twin pairs at age 9, and 5,716 twin pairs at age 12. Reduced sample size at age 9 can be explained by a shift in contacting scheme from phone to in-person interviews.
Statistical Analysis
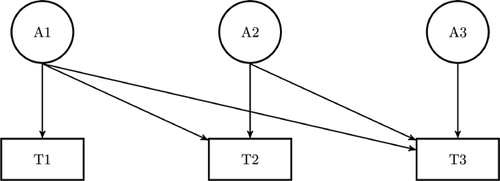
To gain a first impression of stability and heritability of aggression across age, longitudinal within person correlations, twin correlations, and cross-twin-cross-age correlations were estimated for each cohort as a function of zygosity. Next, we applied longitudinal analyses to investigate to what extent preexisting and new genetic and environmental factors influence the dynamic development of aggression. Data from three time points (T1, T2, T3) in the two studies were analyzed, corresponding in NTR to data from ages 7, 9/10, and 12 years and in the TEDS cohort to ages 7, 9, and 12 years. A Cholesky decomposition was fitted to the raw data in which subsequent levels of problem behavior are influenced by latent variables (additive genetic, common environmental, and unique environmental variables) of the current as well as all prior ages (Fig. 1). Hence, for each study (NTR and TEDS) and sex (males and females) the covariance matrix is expressed as a composition of 18 parameters (six parameters from the A, C, and E variables to the phenotype, respectively). Using this decomposition, one can decompose variance into effects due to current and prior latent variables [Franić et al., 2014]. The significance of common environmental influences was tested by dropping the C effect from the model, and evaluating the drop in goodness of fit of the model. Sex-differences in parameter estimates (effects of A, C, and E) were tested by comparing models that allowed for sex differences in variance components to models that constrained these estimates to be the same, as well as to a scalar model in which male standardized variance components are the same as those in females, but allowing for variance differences between the sexes. Similarly, we tested the equality of heritability across the two studies, by constraining estimates of variance components of TEDS to be proportional to those of NTR by allowing for differences in unstandardized variance components between the two studies but constraining standardized components to be the same. All models allowed for mean differences between boys and girls and between different ages. Model comparisons were done by likelihood ratio tests. Parameter estimation was based on raw-data maximum likelihood in the open resource software OpenMx [Boker et al., 2011]. Based on the best fitting model, we estimated the genetic and environmental influences on the variance of aggression at each age and the covariance between ages. In addition, we calculated the genetic and environmental correlations.
RESULTS
Descriptives
The sex distribution in both cohorts was 49% male and 51% female. Aggression scores slightly declined in boys and girls when they grow up from age 7 to12 years. Figure 2 shows the mean aggression scores of CBCL and SDQ. Boys scored higher than girls, but for both scales, the sex differences attenuated with age. Variance is larger for boys than girls for both cohorts at all ages. Variances in MZ and DZ twins are very similar, ruling out important contributions of sibling interaction or rater contrast effects (Table I) [Eaves et al., 1978; Boomsma, 2014]. The longitudinal phenotypic correlations in boys and in girls (see Table II) are high and reflect strong stability of aggressive behavior between ages 7 and 12. The twin correlations are consistently higher for MZ than DZ pairs suggesting additive genetic influences on aggression regardless of assessment instrument (see diagonal in Table III). Correlations between same-sex and opposite-sex DZ twin pairs are similar, indicating no qualitative sex-differences. In addition, twin correlations seem similar across different age groups indicating comparable genetic influences throughout development. Genetic influences on the covariance of aggression between ages is also to be expected given the higher MZ than DZ cross-twin-cross-age correlations (see off-diagonal estimates in Table III).
| Measurement | Age | Variance | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cohort | n | Mean | Sd | Median | Range | MZM | DZM | MZF | DZF | DOSmf | DOSfm |
| NTR | |||||||||||
| 7 | 21530 | 23.12 | 4.91 | 7.35 | 6.17–9.88 | 27.83 | 29.20 | 20.15 | 19.09 | 24.28 | 21.85 |
| 9/10 | 17114 | 22.60 | 4.82 | 9.96 | 8.71–13.88 | 28.71 | 28.82 | 17.56 | 19.40 | 22.58 | 21.00 |
| 12 | 14352 | 21.81 | 4.35 | 12.17 | 11.02–15.41 | 21.65 | 23.30 | 15.56 | 14.86 | 20.24 | 17.24 |
| TEDS | |||||||||||
| 7 | 13794 | 1.68 | 1.62 | 7.04 | 5.57–8.62 | 2.86 | 3.08 | 2.15 | 2.39 | 2.63 | 2.65 |
| 9 | 6056 | 1.27 | 1.44 | 9.01 | 8.08–11.34 | 2.46 | 2.38 | 1.75 | 1.80 | 1.76 | 2.29 |
| 12 | 11432 | 1.32 | 1.46 | 11.44 | 9.79–14.35 | 2.19 | 2.48 | 1.78 | 2.00 | 2.26 | 2.06 |
- Twins are categorized into either male or female monozygotic (MZM, MZF), male or female same-sex dizygotic (DZM, DZF), or opposite-sex dizygotic twin pairs as either female-male or male-female (DOSDZfm, DOSDZmf).
| NTR | 7 years | 9/10 years | 12 years |
|---|---|---|---|
| 7 years | 0.70 (0.69, 0.71) | 0.61 (0.59, 0.63) | |
| 9/10 years | 0.71 (0.7, 0.73) | 0.69 (0.67, 0.7) | |
| 12 years | 0.63 (0.61, 0.65) | 0.74 (0.72, 0.75) |
| TEDS | 7 years | 9 years | 12 years |
|---|---|---|---|
| 7 years | 0.55 (0.52, 0.58) | 0.5 (0.48, 0.53) | |
| 9 years | 0.58 (0.55, 0.61) | 0.55 (0.52, 0.58) | |
| 12 years | 0.55 (0.53, 0.57) | 0.62 (0.59, 0.65) |
- The 95% confidence intervals are given in parenthesis.
| NTR | 7 years | 9/10 years | 12 years |
|---|---|---|---|
| MZ | |||
| 7 years | 0.84a (0.82, 0.85)/0.79b (0.78, 0.81) | 0.65 (0.63, 0.68) | 0.56 (0.53, 0.59) |
| 9/10 years | 0.59 (0.57, 0.62) | 0.83a (0.82, 0.85)/0.77b (0.75, 0.78) | 0.65 (0.63, 0.67) |
| 12 years | 0.54 (0.51, 0.57) | 0.56 (0.54, 0.59) | 0.81a (0.79, 0.82)/0.78b (0.76, 0.79) |
| DZ | |||
| 7 years | 0.51a (0.48, 0.54)/0.44b (0.41, 0.47) | 0.39 (0.36, 0.42) | 0.34 (0.31, 0.37) |
| 9/10 years | 0.37 (0.33, 0.4) | 0.47a (0.44, 0.5)/0.44b (0.41, 0.47) | 0.35 (0.31, 0.38) |
| 12 years | 0.33 (0.29, 0.37) | 0.36 (0.32, 0.39) | 0.46a (0.43, 0.49)/0.47b (0.44, 0.5) |
| DOS | |||
| 7 years | 0.44c (0.41, 0.47)/0.46d (0.43, 0.49) | 0.32 (0.29, 0.36) | 0.35 (0.31, 0.38) |
| 9/10 years | 0.32 (0.28, 0.35) | 0.39c (0.36, 0.42)/0.44d (0.41, 0.47) | 0.36 (0.33, 0.4) |
| 12 years | 0.29 (0.25, 0.32) | 0.29 (0.25, 0.32) | 0.53c (0.5, 0.56)/0.46d (0.43, 0.49) |
| TEDS | 7 years | 9 years | 12 years |
|---|---|---|---|
| MZ | |||
| 7 years | 0.76a (0.74, 0.78)/0.72b (0.7, 0.74) | 0.45 (0.41, 0.48) | 0.5 (0.46, 0.53) |
| 9 years | 0.41 (0.37, 0.44) | 0.8a (0.78, 0.81)/0.76b (0.74, 0.78) | 0.5 (0.47, 0.53) |
| 12 years | 0.44 (0.41, 0.47) | 0.48 (0.45, 0.52) | 0.74a (0.72, 0.76)/0.78b (0.76, 0.8) |
| DZ | |||
| 7 years | 0.47a (0.43, 0.5)/0.44b (0.4, 0.47) | 0.32 (0.29, 0.36) | 0.32 (0.28, 0.35) |
| 9 years | 0.28 (0.24, 0.32) | 0.49a (0.45, 0.52)/0.59b (0.56, 0.62) | 0.29 (0.25, 0.32) |
| 12 years | 0.24 (0.2, 0.28) | 0.3 (0.26, 0.34) | 0.46a (0.42, 0.49)/0.52b (0.49, 0.55) |
| DOS | |||
| 7 years | 0.46c (0.43, 0.49)/0.42d (0.39, 0.46) | 0.2 (0.16, 0.24) | 0.29 (0.25, 0.33) |
| 9 years | 0.23 (0.19, 0.27) | 0.52c (0.49, 0.55)/0.45d (0.42, 0.49) | 0.35 (0.31, 0.38) |
| 12 years | 0.26 (0.22, 0.29) | 0.29 (0.25, 0.33) | 0.52c (0.49, 0.55)/0.5d (0.47, 0.53) |
- Twins are categorized into either monozygotic (MZ), same-sex dizygotic (DZ), or other-sex dizygotic twin pairs (DOS). Within each zygosity table the upper triangle displays the correlation of twin pairs in which the older twin is male, the lower-triangle represents correlation of twin pairs in which the older twin is female. The 95% confidence intervals are given in parentheses.
- a male.
- b female.
- c First twin is male.
- d First twin is female.
Longitudinal Genetic Modelling
The twin and cross-twin-cross age correlations within NTR and TEDS suggest stable genetic influences on aggression and genetic influences on the stability of aggression. To investigate the genetic architecture in more detail, a genetic Cholesky decomposition was applied to the raw data in the two studies. For model comparison we considered the fully saturated genetic model as a reference to test for the significance of common environment (C) shared by twins from the same family, and quantitative sex differences in parameter estimates. In addition we also investigated a sex specific scalar model, in which we constrained variance components of male and female twins to be proportional to each other; that is, we specified equal standardized variance components between the sexes while allowing for differences in unstandardized variances components. Table IV presents the model fitting results and indicates significant effects for the common environmental component in both studies (NTR: χ212 = 127.77, P < 0.0001 TEDS: χ212 = 177.28, P < 0.0001). Constraining parameters to be the same for boys and girls (NTR: χ218 = 596.82, P < 0.0001; TEDS: χ218 = 193.12, P < 0.0001), as well as constraining male variance components proportional to female estimates (NTR: χ215 = 101.63, P < 0.0001; TEDS: χ215 = 45.15, P < 0.0001) resulted in a significant worsening of model fit, although the differences are small. In order to test for similarities in genetic architecture of aggression as assessed with the CBCL and the SDQ we tested for equal standardized components by constraining parameters of TEDS to be proportional to those in NTR. This resulted in a significantly worse fit compared to study-specific estimates (χ230= 435.07, P < 0.0001). Thus, there are significant differences between the genetic architecture of aggression based on the CBCL (NTR) and the SDQ (TEDS). The differences are small, as may be seen from the study specific variance and covariance components for NTR and TEDS in Table V. The heritability of aggression at ages 7, 9/10, and 12 years ranges between 42% and 78%. The lowest estimate (42%) is observed for females at age 10 in the TEDS sample, while the highest estimates are observed for females at age 7 (78%) and 10 (76%) in the NTR sample. A number of differences could be observed between NTR and TEDS. Heritability is somewhat lower in TEDS than NTR across sexes and differences between boy and girl estimates are slightly larger in NTR. Nevertheless, overall both studies are rather similar with respect to the influence of genetic and environmental components on aggressive behavior, despite the differences in assessment instruments. Partitioning of genetic effects into influences from prior ages demonstrates that the latent genetic factor in T1 is a major contributor to the genetic variance at T2 and T3, indicating that preexisting genetic factors play an increasingly important role in explaining variation in aggressive behavior (see Table VI). The influence of T1 on subsequent ages is stronger in NTR than TEDS, which is also reflected by the relatively larger longitudinal correlation in NTR. This stability in underlying genetic effects is also reflected in the high genetic correlations (Table VII). Genetic correlations are in the ranges of 0.76–0.85 for NTR and 0.64 and 0.77 for TEDS. These high genetic correlation in combination with the significant genetic influences on the stability of aggressive behavior indicates that genes are the major driving force of the persistence of aggressive behavior throughout childhood regardless of the assessment instrument.
| Reference | Model | Parameters | −2ll | df | AIC | Δ−2ll | Δdf | P |
|---|---|---|---|---|---|---|---|---|
| NTR | ||||||||
| Full cholesky | Cholesky: sex moderated AE | 30 | 297589.30 | 55098 | 187393.30 | 127.77 | 12 | <0.0001 |
| Full cholesky | Cholesky: no-sex moderated ACE | 24 | 298058.36 | 55104 | 187850.36 | 596.83 | 18 | <0.0001 |
| Full cholesky | Cholesky: scaled sex ACE | 27 | 297563.15 | 55101 | 187361.15 | 101.63 | 15 | <0.0001 |
| TEDS | ||||||||
| Full cholesky | Cholesky: sex moderated AE | 30 | 101529.26 | 30983 | 39563.26 | 177.29 | 12 | <0.0001 |
| Full cholesky | Cholesky: no-sex moderated ACE | 24 | 101545.10 | 30989 | 39567.10 | 193.13 | 18 | <0.0001 |
| Full cholesky | Cholesky: scaled sex ACE | 27 | 101397.13 | 30986 | 39425.13 | 45.16 | 15 | <0.0001 |
| Combined | ||||||||
| Cholesky: study specific | 84 | 398813.5 | 86057 | 226699.5 | ||||
| Cholesky: study specific | Cholesky: combined estimates | 54 | 399248.6 | 86087 | 227074.6 | 435.073 | 30 | <0.0001 |
| A | C | E | |||||||
|---|---|---|---|---|---|---|---|---|---|
| NTR | 7 years | 9/10 years | 12 years | 7 years | 9/10 years | 12 years | 7 years | 9/10 years | 12 years |
| 7 years | 19.4 (17.85, 21.1)/15.39 (14.36, 16.23) | 15.03 (13.76, 16.34) | 13.76 (12.63, 14.87) | 5.48 (3.76, 7.14)/0.22 (0, 0.93) | 4.01 (2.53, 5.45) | 1.87 (0.72, 3.12) | 4.67 (4.38, 4.99)/4.13 (3.89, 4.38) | 1.89 (1.6, 2.18) | 1.05 (0.75, 1.36) |
| 9/10 years | 11.14 (10.26, 11.87) | 19.72 (18.21, 21.33)/14.05 (13.03, 14.91) | 15.24 (14.05, 16.43) | 0.17 (−0.01, 0.76) | 4.47 (2.73, 6.2)/0.18 (0, 0.84) | 2.27 (1.06, 3.59) | 2.1 (1.88, 2.34) | 4.79 (4.47, 5.15)/4.32 (4.04, 4.63) | 1.86 (1.57, 2.16) |
| 12 years | 9.25 (8.44, 9.95) | 9.71 (8.85, 10.48) | 16.41 (15.02, 17.75)/9.64 (8.65, 10.65) | 0.28 (0, 0.85) | 0.34 (−0.38, 0.99) | 3.1 (1.94, 4.47)/2.42 (1.59, 3.28) | 1.16 (0.96, 1.38) | 1.65 (1.42, 1.89) | 4.31 (3.98, 4.67)/3.51 (3.26, 3.78) |
| 7 years | 0.66 (0.6, 0.71)/0.78 (0.74, 0.8) | 0.72 (0.65, 0.79) | 0.82 (0.75, 0.9) | 0.19 (0.13, 0.24)/0.01 (0, 0.05) | 0.19 (0.12, 0.25) | 0.11 (0.04, 0.18) | 0.16 (0.15, 0.17)/0.21 (0.2, 0.22) | 0.09 (0.08, 0.1) | 0.06 (0.05, 0.08) |
| 9/10 years | 0.83 (0.78, 0.86) | 0.68 (0.63, 0.74)/0.76 (0.72, 0.78) | 0.79 (0.72, 0.85) | 0.01 (0, 0.06) | 0.15 (0.1, 0.21)/0.01 (0, 0.04) | 0.12 (0.06, 0.18) | 0.16 (0.14, 0.18) | 0.17 (0.15, 0.18)/0.23 (0.22, 0.25) | 0.1 (0.08, 0.11) |
| 12 years | 0.87 (0.81, 0.9) | 0.83 (0.77, 0.9) | 0.69 (0.63, 0.74)/0.62 (0.56, 0.68) | 0.03 (0, 0.08) | 0.03 (−0.03, 0.08) | 0.13 (0.08, 0.18)/0.16 (0.1, 0.21) | 0.11 (0.09, 0.13) | 0.14 (0.12, 0.16) | 0.18 (0.17, 0.2)/0.23 (0.21, 0.24) |
| TEDS | 7 years | 9 years | 12 years | 7 years | 9 years | 12 years | 7 years | 9 years | 12 years |
|---|---|---|---|---|---|---|---|---|---|
| 7 years | 1.88 (1.64, 2.16)/1.44 (1.22, 1.63) | 1.06 (0.83, 1.37) | 1.1 (0.94, 1.28) | 0.44 (0.18, 0.67)/0.24 (0.09, 0.45) | 0.3 (0.03, 0.56) | 0.24 (0.09, 0.41) | 0.69 (0.64, 0.75)/0.61 (0.56, 0.66) | 0.21 (0.14, 0.27) | 0.15 (0.11, 0.2) |
| 9 years | 0.71 (0.57, 0.86) | 1.45 (1.18, 1.74)/0.78 (0.63, 0.94) | 0.98 (0.81, 1.19) | 0.21 (0.08, 0.37) | 0.49 (0.25, 0.79)/0.65 (0.48, 0.83) | 0.32 (0.14, 0.48) | 0.2 (0.16, 0.23) | 0.53 (0.47, 0.6)/0.4 (0.36, 0.45) | 0.23 (0.18, 0.29) |
| 12 years | 0.83 (0.7, 0.96) | 0.71 (0.58, 0.85) | 1.4 (1.21, 1.6)/1.08 (0.94, 1.23) | 0.16 (0.06, 0.29) | 0.21 (0.08, 0.35) | 0.47 (0.3, 0.65)/0.47 (0.34, 0.62) | 0.07 (0.04, 0.11) | 0.13 (0.09, 0.16) | 0.58 (0.53, 0.64)/0.4 (0.37, 0.43) |
| 7 years | 0.62 (0.55, 0.71)/0.63 (0.53, 0.7) | 0.67 (0.51, 0.87) | 0.74 (0.62, 0.85) | 0.14 (0.06, 0.22)/0.1 (0.04, 0.19) | 0.19 (0.02, 0.34) | 0.16 (0.06, 0.26) | 0.23 (0.21, 0.25)/0.27 (0.24, 0.29) | 0.13 (0.09, 0.17) | 0.1 (0.07, 0.13) |
| 9 years | 0.63 (0.5, 0.76) | 0.59 (0.47, 0.69)/0.42 (0.34, 0.52) | 0.64 (0.53, 0.77) | 0.19 (0.07, 0.32) | 0.2 (0.1, 0.31)/0.36 (0.27, 0.44) | 0.21 (0.09, 0.31) | 0.17 (0.14, 0.21) | 0.21 (0.19, 0.25)/0.22 (0.2, 0.25) | 0.15 (0.12, 0.19) |
| 12 years | 0.78 (0.66, 0.89) | 0.68 (0.55, 0.81) | 0.57 (0.5, 0.65)0.55 (0.48, 0.63) | 0.15 (0.05, 0.26) | 0.21 (0.08, 0.32) | 0.19 (0.12, 0.26)/0.24 (0.17, 0.31) | 0.07 (0.04, 0.1) | 0.12 (0.09, 0.15) | 0.24 (0.22, 0.26)/0.2 (0.19, 0.22) |
- Diagonal; first number is for males, second for females. Off-diagonal; below is for males, above is for females. The 95% confidence intervals are given in parentheses.
| Male | Female | |||||
|---|---|---|---|---|---|---|
| Cohort | A | C | E | A | C | E |
| NTR | ||||||
| 7 years | 0.66 (0.6, 0.71) | 0.19 (0.13, 0.24) | 0.16 (0.15, 0.17) | 0.78 (0.74, 0.8) | 0.01 (0, 0.05) | 0.21 (0.2, 0.22) |
| 9/10 years | 0.68 (0.63, 0.74) [0.4, 0.28] | 0.15 (0.1, 0.21) [0.1, 0.05] | 0.17 (0.15, 0.18) [0.03, 0.14] | 0.76 (0.72, 0.78) [0.43, 0.33] | 0.01 (0, 0.04) [0.01, 0] | 0.23 (0.22, 0.25) [0.06, 0.17] |
| 12 years | 0.69 (0.63, 0.74) [0.41, 0.11, 0.17] | 0.13 (0.08, 0.18) [0.03, 0.02, 0.08] | 0.18 (0.17, 0.2) [0.01, 0.02, 0.15] | 0.62 (0.56, 0.68) [0.36, 0.1, 0.16] | 0.16 (0.1, 0.21) [0.02, 0.02, 0.12] | 0.23 (0.21, 0.24) [0.03, 0.02, 0.18] |
| TEDS | ||||||
| 7 years | 0.62 (0.55, 0.71) | 0.14 (0.06, 0.22) | 0.23 (0.21, 0.25) | 0.63 (0.53, 0.7) | 0.1 (0.04, 0.19) | 0.27 (0.24, 0.29) |
| 9 years | 0.59 (0.47, 0.69) [0.24, 0.35] | 0.2 (0.1, 0.31) [0.08, 0.12] | 0.21 (0.19, 0.25) [0.02, 0.19] | 0.42 (0.34, 0.52) [0.19, 0.23] | 0.36 (0.27, 0.44) [0.1, 0.25] | 0.22 (0.2, 0.25) [0.03, 0.17] |
| 12 years | 0.57 (0.5, 0.65) [0.26, 0.07, 0.24] | 0.19 (0.12, 0.26) [0.06, 0.03, 0.1] | 0.24 (0.22, 0.26) [0.01, 0.04, 0.19] | 0.55 (0.48, 0.63) [0.25, 0.1, 0.2] | 0.24 (0.17, 0.31) [0.05, 0.01, 0.18] | 0.2 (0.19, 0.22) [0, 0.02, 0.18] |
- Variance component are partitioned into effects due to effects from different time points (brackets). The first, second and third number are effects due to first, second or third genetic/environmental factors, respectively. Confidence intervals (95%) for the standardized variance component are given in parentheses.
| NTR | 7 years | 9/10 years | 12 years |
|---|---|---|---|
| 7 years | 0.76 (0.73, 0.78) | 0.76 (0.72, 0.8) | |
| 9/10 years | 0.77 (0.72, 0.8) | 0.83 (0.79, 0.88) | |
| 12 years | 0.77 (0.73, 0.81) | 0.85 (0.81, 0.89) |
| TEDS | 7 years | 9 years | 12 years |
|---|---|---|---|
| 7 years | 0.67 (0.57, 0.76) | 0.67 (0.58, 0.75) | |
| 9 years | 0.64 (0.54, 0.76) | 0.77 (0.68, 0.85) | |
| 12 years | 0.68 (0.6, 0.76) | 0.69 (0.61, 0.78) |
- Upper triangle displays female and lower male genetic correlation. The 95% confidence intervals are given in parentheses.
DISCUSSION
In the current, paper we analyzed data from two large longitudinal cohorts with information on childhood aggression in twins to investigate the underlying sources of individual differences and stability of this trait. A longitudinal twin model of aggression data assessed with the commonly used CBCL and SDQ reveals that genetic factors are not only the largest contributors to individual differences in aggression at each age, but also the major contributor to stability of aggressive behavior throughout childhood. We observed significant sex differences, mainly regarding the influence of the common environment, as well as significant differences in estimates between the two studies. However, careful inspection of estimates suggests that these difference between results based on the CBCL (18 items) and the SDQ (5 items) for age specific source of variation as well as phenotypic stability and its underlying sources are rather minor.
Some limitations should be taken into account while interpreting the results of this study. First, the analyses are based on maternal ratings of overall aggression. Previous research has shown that about 20% of the variance in maternal ratings during childhood is accounted for by rater bias [Bartels et al., 2007]. The study by Haberstick et al. [2006] also demonstrated considerable heritability differences between maternal and teacher ratings (maternal: 76–84%; teacher: 42–62%), with higher non-shared environmental influence in teacher ratings, In addition maternal-teacher correlations are low, suggesting situation specific influences. The study by Hudziak et al. [2003], though, concludes that, despite the differences in informants, reflected by the lack of correlation often seen among maternal, paternal, and teacher reports, similar magnitude of genetic influences for parental and teacher ratings. The two sources of information, that is, parental and teacher ratings, also led to some differences. Analyses of maternal and paternal CBCL reports provide consistent evidence of additive genetic and shared and unique environmental influences across development, while teacher reports provide no evidence of shared environment.
Furthermore, the focus on overall aggression ignores the differences in genetic architecture that may exist for subtypes of aggression. For example, Ligthart et al. [2005] identified two aggression subtypes (relational and direct aggression) within the aggression syndrome scale of the CBCL and report that both were influenced by one underlying set of shared environmental factors, but only partly by the same genes (the genetic correlation was 0.54 for boys and 0.43 for girls). Ghodsian-Carpey and Baker [1987] showed that surveys which measure more subtle forms of aggression (such as teasing and noncompliance) seem to lead to lower heritability estimates than surveys assessing more extreme forms of aggression (such as destructiveness and insult).
Overall, the results of our analyses generally agree with previous longitudinal studies on aggression. In this large study, we confirm the absence of qualitative sex-differences. Based on the model fitting results, we also conclude that there are significant quantitative sex-differences. Careful observation of the unstandardized and standardized variance components reveals that for NTR this effect is mainly driven by the presence of shared environmental influences in males at younger ages. Furthermore, the influences of non-shared environmental effects are larger for girls than for boys. For TEDS the quantitative sex-differences are less pronounced. Quantitative sex-differences in heritability in our study with large sample sizes were small. Previous studies, which reported sex-differences in heritability, tended to show a diminishing effect by age, similar to our results. The absence of qualitative sex-differences, the relative small quantitative sex-differences, and the comparable genetic architecture of aggression throughout childhood based on two different assessment instruments is welcome news for large scale gene-finding studies. Given that sample size is one of the major factors in these studies, results that the same genes with similar effect size might be of importance for boys and girls, for different age groups, for stability of aggression, and for aggressive behavior based on different assessment instruments enables worldwide collaborative projects for GWA studies.
From a clinical point of view, the stability of aggression and the stable influences of genetic factors from young age onwards indicates that a wait and see policy might not be the best approach to helping children and their families who suffer from aggressive problems. Detection and identification of aggressive problems at a young age might help to prevent further suffering. Common environmental influences shared by children from the same family are significant in boys in both the NTR and TEDS, but in girls only in TEDS and not NTR. This is an interesting finding which may point to cultural differences between the Netherlands and the UK, but may also result from the different instruments used in the two studies. A longitudinal study of US-American twins aged 7–12 years old using CBCL demonstrated similar genetic correlations and parameter estimates as the here presented results in NTR [Haberstick et al., 2006], suggesting that differences between the two studies might be driven by the used instruments. We need to investigate whether some instruments may be more sensitive to detecting influences of the shared family environment than others and also whether the same instrument behaves differently across cultures. For both NTR and TEDS non-shared environmental influences are stable and age specific. While this can be partly explained by measurement errors an additional possiblity is that the effect of environmental events on aggressive behavior is temporary and decays over time. Suggesting constant but heterogeneous environmental effects. Taken together, these results, in combination with the relatively high heritability estimates, plea for studies into gene-environment interplay to inform the development of new treatment strategies to target aggressive behavior in children.
Based on two large longitudinal samples of twin data from the UK and the Netherlands we conclude that childhood aggression is a stable trait. Individual differences at the various ages are mainly accounted for by genetic differences between individuals. Additionally, genetic influences are also found to be the major source of stability in aggressive behavior throughout childhood. Based on the large sample size, we can furthermore conclude that shared environmental influences are significant, especially for boys. The picture for girls is less clear since inconsistent findings are observed for the NTR versus the TEDS sample.




