Association analyses between brain-expressed fatty-acid binding protein (FABP) genes and schizophrenia and bipolar disorder†
How to Cite this Article: Iwayama Y, Hattori E, Maekawa M, Yamada K, Toyota T, Ohnishi T, Iwata Y, Tsuchiya KJ, Sugihara G, Kikuchi M, Hashimoto K, Iyo M, Inada T, Kunugi H, Ozaki N, Iwata N, Nanko S, Iwamoto K, Okazaki Y, Kato T, Yoshikawa T. 2009. Association Analyses Between Brain-Expressed Fatty-Acid Binding Protein (FABP) Genes and Schizophrenia and Bipolar Disorder. Am J Med Genet Part B 153B:484–493.
Abstract
Deficits in prepulse inhibition (PPI) are a biological marker for psychiatric illnesses such as schizophrenia and bipolar disorder. To unravel PPI-controlling mechanisms, we previously performed quantitative trait loci (QTL) analysis in mice, and identified Fabp7, that encodes a brain-type fatty acid binding protein (Fabp), as a causative gene. In that study, human FABP7 showed genetic association with schizophrenia. FABPs constitute a gene family, of which members FABP5 and FABP3 are also expressed in the brain. These FABP proteins are molecular chaperons for polyunsaturated fatty acids (PUFAs) such as arachidonic and docosahexaenoic acids. Additionally, the involvement of PUFAs has been documented in the pathophysiology of schizophrenia and mood disorders. Therefore in this study, we examined the genetic roles of FABP5 and 3 in schizophrenia (N = 1,900 in combination with controls) and FABP7, 5, and 3 in bipolar disorder (N = 1,762 in the case–control set). Three single nucleotide polymorphisms (SNPs) from FABP7 showed nominal association with bipolar disorder, and haplotypes of the same gene showed empirical associations with bipolar disorder even after correction of multiple testing. We could not perform association studies on FABP5, due to the lack of informative SNPs. FABP3 displayed no association with either disease. Each FABP is relatively small and it is assumed that there are multiple regulatory elements that control gene expression. Therefore, future identification of unknown regulatory elements will be necessary to make a more detailed analysis of their genetic contribution to mental illnesses. © 2009 Wiley-Liss, Inc.
INTRODUCTION
Despite entering the era of whole genome association analyses, the unequivocal identification of susceptibility genes for schizophrenia and bipolar disorder still warrants further work [Wellcome Trust Consortium, 2007; Baum et al., 2008; O'Donovan et al., 2008; Sklar et al., 2008; Hattori et al., 2009; Need et al., 2009]. One of the reasons for this may be that current diagnostic categorization is largely dependent on the subjective evaluation of patients' feelings and state of mood. This may result in etiologically (biologically) extremely heterogeneous disease states being categorized together [Need et al., 2009]. As an alternative approach, the analysis of biological traits associated with psychiatric illnesses called “endophenotypes” has gained importance. Although endophenotypes are an idealized concept, they are expected to assist in deconstructing complex diseases, allowing for easier genetic analyses [Gottesman and Gould, 2003; Gur et al., 2007].
As an example of an endophenotype, deficits in prepulse inhibition (PPI) have been well documented in psychiatric illnesses including schizophrenia and bipolar disorder [Braff et al., 2001; Giakoumaki et al., 2007]. The experimental advantage of PPI is that it is evaluable in animals. To identify the genes that control PPI, we performed quantitative trait loci analysis in mice, and detected a gene encoding Fabp7 (fatty acid binding protein 7, brain type) as a causative genetic substrate [Watanabe et al., 2007]. Furthermore, the human orthologue FABP7 (located on chromosome 6q22.31) was associated with schizophrenia [Watanabe et al., 2007]. The FABPs constitute a gene family and at least 12 members have been reported [for review see Liu et al., 2008; Furuhashi and Hotamisligil, 2008]. Brain-expressed FABPs include FABP5 (chromosome 8q21.13) and FABP3 (chromosome 1p35.2), along with FABP7 [Owada, 2008]. FABP proteins are lipid chaperons, and the ligands for the brain-expressed FABPs are thought to be polyunsaturated fatty acids (PUFAs) such as arachidonic (AA) and docosahexaenoic acid (DHA) [Furuhashi and Hotamisligil, 2008].
Accumulating evidence suggests roles for PUFAs in both schizophrenia and mood disorders [for review see Richardson, 2004]. Therefore in this study, we set out to expand our prior genetic association analysis (that is between FABP7 and schizophrenia [Watanabe et al., 2007]), to between FABPs 5 and 3 and schizophrenia and between FABPs 7, 5, and 3 and bipolar disorder.
MATERIALS AND METHODS
Subjects
The set of schizophrenia and age-/sex-matched control samples consisted of 950 unrelated patients with schizophrenia (447 men, 503 women; mean age 47.0 ± 13.7 years) and controls (447 men, 503 women; mean age 46.9 ± 13.6 years). The sample panel for the bipolar study was the same as used in the COSMO consortium study [Ohnishi et al., 2007], which comprises 867 unrelated bipolar patients (425 men, 442 women; mean age 50.7 ± 14.2 years) and 895 age- and sex-matched controls (445 men, 450 women; mean age 49.9 ± 13.5 years). All samples are of Japanese origin. In our previous genome-wide analysis of a sample set consisting of subjects recruited at almost the same geographical locations as the bipolar case–control set in the current study, little effect of population stratification was detected by principal components analysis [Hattori et al., 2009] and this finding was consistent with another recent report [Yamaguchi-Kabata et al., 2008]. While bipolar case–control recruitment was spread over the Hondo area in Japan, schizophrenia case–control recruitment was restricted to the Kanto district, which includes Tokyo and its surrounding areas, and overlaps to a limited extent with Hondo. Therefore, population stratification should be negligible. All patients had a consensual diagnosis of schizophrenia or bipolar disorder according to DSM-IV criteria, from at least two experienced psychiatrists. Control subjects were recruited from hospital staff and volunteers who showed no present or past evidence of psychoses, during brief interviews by psychiatrists. The current study was approved by the Ethics Committees of all participating institutes. All participants provided written informed consent.
Re-Sequencing Analyses of FABP7 and FABP5
We previously performed a genetic association study between schizophrenia and FABP7 (at chr6: 123142345–123146917 using the UCSC database: http://genome.ucsc.edu/cgi-bin/hgGateway?org=Human&db=hg18&hgsid=121236003), and reported nominal association of a missense polymorphism [rs2279381; 182C > T (Thr61Met) (F06 in Fig. 1)] and its spanning haplotype with schizophrenia [Watanabe et al., 2007]. Assuming the possibility of additional functional SNPs (to Thr61Met) we re-sequenced the entire gene region (spanning 908 bp upstream of exon 1 to 347 bp downstream of exon 4: total length 5,826 bp) using 10 randomly chosen patients with schizophrenia and 10 bipolar disorder samples. Information on the primer sets and PCR conditions for this analysis is available upon request. Sequencing was performed using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) and the ABI PRISM 3730 Genetic Analyzer (Applied Biosystems). Polymorphisms were detected by the SEQUENCHER program (Gene Codes Corporation, Ann Arbor, MI).
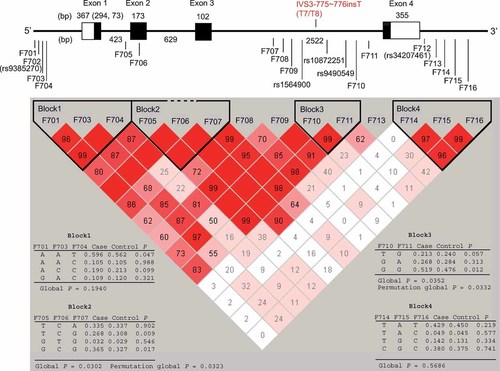
Genomic structure, polymorphic sites and LD block structure of the FABP7 gene. In the upper panel, exons are denoted as boxes, with coding regions in black and 5′-/3′-untranslated regions in white. The sizes of each exon and intron are also shown. In the lower panel, the number in each cell represents the LD parameter D′ (×100), blank cells mean D′ = 1. Each cell is painted in a graduated color relative to the strength of LD between markers, which is defined by both the D′ value and confidence bounds on D′. The results of block-based haplotype analysis in bipolar disorder are also shown for LD blocks 1 through 4, along with haplotype frequencies and global P values.
For analysis of FABP5 (at chr8: 82355340–82359563 on the UCSC database), since there are no SNPs in the HapMap database for the Japanese population (rel #23a) (http://www.hapmap.org/index.html.ja), we re-sequenced the gene region (spanning 897 bp upstream of exon 1 to 447 bp downstream of exon 4: total length 5,568 bp) using the same 10 schizophrenic and 10 bipolar samples described previously.
This sample set used for the mutation screen will fail to detect a variant if all the cases with bipolar disorder and schizophrenia are either homozygous for a risk allele or for a non-risk allele. This is unlikely to be the case for common variations. The current sample set, which consists of 20 cases and no controls, provides a sensitivity of >0.99 for a risk allele, with a frequency range of 0.1–0.87. This is under the assumption of Hardy-Weinberg equilibrium in the general population and a multiplicative model with a genotype relative risk of 1.2.
Information on the primer sets and PCR conditions for this analysis is available upon request.
SNP Selection and Genotyping
For FABP7, we selected tag SNPs from all SNPs detected by re-sequencing, and from SNPs located from the 10 kb up- and down-stream regions of the gene [the HapMap data for the Japanese population (rel #23a)]. Tag SNPs were selected by Carlson's greedy algorithm, which is implemented in the LdSelect program [Carlson et al., 2004]. The minor allele frequency and the r2 threshold were set to 0.1 and 0.85, respectively. The same tag SNP selection criteria were applied to FABP3.
SNP genotyping was performed using the TaqMan system (Applied Biosystems, Foster City, CA) according to the recommendations of the manufacturer. PCR was performed using an ABI 9700 thermocycler and fluorescent signals were analyzed using an ABI 7900 sequence detector single point measurement and SDS v2.3 software (Applied Biosystems).
Copy Number Polymorphism (CNP) Analysis of FABP3
Because the UCSC database (assembly March, 2006) showed a large CNP (cnp20; position: chr1: 31454968–32238918) spanning the entire FABP3 region (at chr1: 31610687–31618510 on the UCSC database), we tested to confirm the existence of CNPs in Japanese subjects using genomic quantitative PCR. The amplicons were set at both the 5′- and 3′-ends of the gene (detailed information is available on request).
Statistical Analyses
Deviations from Hardy–Weinberg equilibrium (HWE) were evaluated by the chi-square test (df = 1). Allele and genotype distributions between patients and controls were compared using Fisher's exact test. To determine the linkage disequilibrium (LD) block structure in each gene region, we used the genotype data from the schizophrenia (cases + controls: N = 1,900) and bipolar disorder sets (cases + controls: N = 1,762) and the Haploview program (http://www.broad.mit.edu/mpg/haploview/) [Barrett et al., 2005].
Haplotype frequency calculations and haplotypic association analyses were performed using the expectation–maximization algorithm implemented in the COCAPHASE program in the UNPHASED v3.0.11 program (http://www.mrc-bsu.cam.ac.uk/personal/frank/software/unphased/) [Dudbridge, 2003].
Statistical power for detecting association was calculated using the Genetic Power Calculator (GPC, http://statgen.iop.kcl.ac.uk/gpc/) [Purcell et al., 2003], under the following parameter assumptions with respect to allelic test statistics: GRR (genetic relative risk) = 1.2, prevalence of disease = 0.01, risk allele frequency = 0.3, α = 0.05 and a multiplicative model of inheritance.
Permutation analysis was performed for correction of multiple testing, using the Haploview software (10,000 runs) [Barrett et al., 2005].
RESULTS
Association Results Between FABP7 and Bipolar Disorder
By re-sequencing analysis of the entire gene region, we detected 12 SNPs (F705–F712, rs1564900, rs10872251, rs9490549, IVS3-775–776InsT in Fig. 1), of which IVS3-775–776insT (T7 or T8: T8 is a minor allele with a frequency of 0.025) was novel. However, there were no new variants that appeared to alter gene function(s). SNPs F01 to F16 (the additional four SNPs are from the HapMap database) were selected as tags, but SNPs F02 (rs9385270) and F712 (rs34207461) could not be typed using the TaqMan method. Accordingly, the remaining 14 SNPs were analyzed.
The allelic and genotypic distributions of each SNP in the bipolar patients and controls are summarized in Table I. All the SNPs were in HWE. SNPs F704 [T allele is over-represented in the bipolar group; OR (95% CI) = 1.15 (1.00–1.31)], F705 [G is over-represented in the bipolar group; OR (95% CI) = 1.20 (1.05–1.38)] and F709 [G is over-represented in the bipolar group; OR (95% CI) = 1.20 (1.05–1.38)] showed nominal associations (P < 0.05). However, after correction by permutation tests, none remained significant. The gene region consisted of four LD blocks (Fig. 1). In haplotype analysis, blocks 2 [T (F705)–C (F706)–G (F707) is over-represented in the control group; OR (95% CI) = 0.82 (0.71–0.95)] [G (F705)–C (F706)–G (F707) is over-represented in the disease group; OR (95% CI) = 1.19 (1.03–1.36)] and 3 [G (F710)–G (F711) is over-represented in the disease group; OR (95% CI) = 1.18 (1.04–1.35)] were associated with disease, even after correction for multiple testing by permutation tests (Fig. 1). The missense SNP F706, previously associated with schizophrenia [Watanabe et al., 2007], was located in block 2. Power analysis gave 72.2% power for the bipolar-control allelic test statistic.
| Our SNP ID and rs# | HWE | N | Allele | P | Genotype | P* | MAF | Permutation P* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | G | A/A | A/G | G/G | ||||||||
| F701 | BP | 0.2267 | 861 | 1532 | 190 | 678 | 176 | 7 | 11.0% | |||
| rs4247671 | CT | 0.3312 | 894 | 1573 | 215 | 0.3693 | 695 | 183 | 16 | 0.2028 | 12.0% | 0.9979 |
| Our SNP ID and rs# | HWE | N | Allele | P | Genotype | P* | MAF | Permutation P* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | C | A/A | A/C | C/C | ||||||||
| F703 | BP | 0.9501 | 865 | 1401 | 329 | 567 | 267 | 31 | 19.0% | |||
| rs12662030 | CT | 0.9158 | 892 | 1404 | 380 | 0.0928 | 553 | 298 | 41 | 0.2393 | 21.3% | 0.8168 |
| Our SNP ID and rs# | HWE | N | Allele | P | Genotype | P* | MAF | Permutation P* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | C | T/T | T/C | C/C | ||||||||
| F704 | BP | 0.1102 | 862 | 1026 | 698 | 294 | 438 | 130 | 40.5% | |||
| rs9372716 | CT | 0.3170 | 893 | 1003 | 783 | 0.0474 | 289 | 425 | 179 | 0.0236 | 43.8% | 0.5500 |
| Our SNP ID and rs# | HWE | N | Allele | P | Genotype | P* | MAF | Permutation P* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | G | T/T | T/G | G/G | ||||||||
| F705 | BP | 0.3365 | 861 | 1037 | 685 | 319 | 399 | 143 | 39.8% | |||
| rs2279382 | CT | 0.4871 | 894 | 1153 | 635 | 0.0099 | 367 | 419 | 108 | 0.0174 | 35.5% | 0.1544 |
| Our SNP ID and rs# | HWE | N | Allele | P | Genotype | P* | MAF | Permutation P* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | C | T/T | T/C | C/C | ||||||||
| F706 (T61M) | BP | 0.3240 | 861 | 56 | 1666 | 0 | 56 | 805 | 3.3% | |||
| rs2279381 | CT | 0.3803 | 895 | 51 | 1739 | 0.4937 | 0 | 51 | 844 | 0.4869 | 2.8% | 0.9998 |
| Our SNP ID and rs# | HWE | N | Allele | P | Genotype | P* | MAF | Permutation P* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | G | A/A | A/G | G/G | ||||||||
| F707 | BP | 0.8253 | 862 | 577 | 1147 | 98 | 381 | 383 | 33.5% | |||
| rs7752838 | CT | 0.2734 | 894 | 603 | 1185 | 0.8864 | 109 | 385 | 400 | 0.8239 | 33.7% | 1.0000 |
| Our SNP ID and rs# | HWE | N | Allele | P | Genotype | P* | MAF | Permutation P* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | C | T/T | T/C | C/C | ||||||||
| F708 | BP | 0.3246 | 862 | 970 | 754 | 280 | 410 | 172 | 43.7% | |||
| rs9401594 | CT | 0.3262 | 895 | 979 | 811 | 0.3594 | 275 | 429 | 191 | 0.6554 | 45.3% | 0.9976 |
| Our SNP ID and rs# | HWE | N | Allele | P | Genotype | P* | MAF | Permutation P* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | G | A/A | A/G | G/G | ||||||||
| F709 | BP | 0.2443 | 857 | 1000 | 714 | 300 | 400 | 157 | 41.7% | |||
| rs9401595 | CT | 0.3465 | 892 | 1120 | 664 | 0.0077 | 345 | 430 | 117 | 0.0093 | 37.2% | 0.1165 |
| Our SNP ID and rs# | HWE | N | Allele | P | Genotype | P* | MAF | Permutation P** | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | G | T/T | T/G | G/G | ||||||||
| F710 | BP | 0.5759 | 858 | 367 | 1349 | 42 | 283 | 533 | 21.4% | |||
| rs9490550 | CT | 0.7948 | 892 | 429 | 1355 | 0.0636 | 53 | 323 | 516 | 0.1713 | 24.0% | 0.6244 |
| Our SNP ID and rs# | HWE | N | Allele | P | Genotype | P* | MAF | Permutation P** | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | G | A/A | A/G | G/G | ||||||||
| F711 | BP | 0.3970 | 859 | 462 | 1256 | 67 | 328 | 464 | 26.9% | |||
| rs9401596 | CT | 0.5371 | 893 | 508 | 1278 | 0.3081 | 76 | 356 | 461 | 0.5911 | 28.4% | 0.9955 |
| Our SNP ID and rs# | HWE | N | Allele | P | Genotype | P* | MAF | Permutation P** | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | C | T/T | T/C | C/C | ||||||||
| F713 | BP | 0.2383 | 859 | 1211 | 507 | 434 | 343 | 82 | 29.5% | |||
| rs9482286 | CT | 0.2359 | 895 | 1283 | 507 | 0.4563 | 467 | 349 | 79 | 0.7505 | 28.3% | 0.9996 |
| Our SNP ID and rs# | HWE | N | Allele | P | Genotype | P* | MAF | Permutation P** | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | C | T/T | T/C | C/C | ||||||||
| F714 | BP | 0.1232 | 858 | 1055 | 661 | 335 | 385 | 138 | 38.5% | |||
| rs6899351 | CT | 0.1382 | 889 | 1107 | 671 | 0.6507 | 355 | 397 | 137 | 0.9025 | 37.7% | 1.0000 |
| Our SNP ID and rs# | HWE | N | Allele | P | Genotype | P* | MAF | Permutation P** | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | G | A/A | A/G | G/G | ||||||||
| F15 | BP | 0.1649 | 856 | 821 | 891 | 207 | 407 | 242 | 48.0% | |||
| rs6919681 | CT | 0.5725 | 893 | 882 | 904 | 0.4168 | 222 | 438 | 233 | 0.5940 | 49.4% | 0.9992 |
| Our SNP ID and rs# | HWE | N | Allele | P* | Genotype | P* | MAF | Permutation P** | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | C | T/T | T/C | C/C | ||||||||
| F716 | BP | 0.7784 | 864 | 746 | 982 | 159 | 428 | 277 | 43.2% | |||
| rs6904500 | CT | 0.7331 | 894 | 810 | 978 | 0.2090 | 186 | 438 | 270 | 0.4068 | 45.3% | 0.9648 |
- BP, bipolar disorder; CT, control; HWE, Hardy–Weinberg equilibrium; MAF, minor allele, frequency.
- Bold P values mean P < 0.05.
- * Evaluated by Fisher's exact test.
- ** Permutation was run 10,000 times.
Re-Sequencing Analysis of FABP5
We screened the gene region (5,568 bp) for polymorphisms using 20 disease samples, and detected a SNP, −36G/C. But the minor allele (C) frequency was 0.025. Therefore, we did not proceed with genetic association studies.
Association Results Between FABP3 and Schizophrenia/Bipolar Disorder
As shown in Figure 2, eight SNPs were selected as tags. LD block analysis showed that SNPs F302–F308 constitute one LD block in both the schizophrenia-control and bipolar disorder-control sample sets (data not shown). None of the 8 SNPs showed association with schizophrenia (Table II) or bipolar disorder (Table III). Also, haplotype analysis showed no association with schizophrenia or bipolar disorder (Table SI). Power analysis gave 75.3% power for the schizophrenia-control allelic test statistic (for the bipolar disorder sample set, see above).
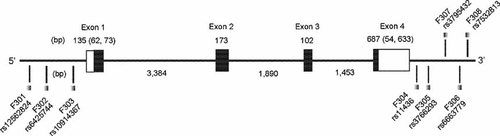
Genomic structure and polymorphic sites in the FABP3 gene. Exons are denoted as boxes, with coding regions in black and 5′-/3′-untranslated regions in white. The sizes of each exon and intron are also shown.
| Our SNP ID and rs# | HWE | N | Allele | P* | Genotype | MAF | P* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| T | C | T/T | T/C | C/C | |||||||
| F301 | SZ | 0.5897 | 942 | 625 | 1259 | 100 | 425 | 417 | 33.2% | ||
| rs12562824 | CT | 0.5250 | 945 | 620 | 1270 | 0.8354 | 106 | 408 | 431 | 32.8% | 0.6886 |
| Our SNP ID and rs# | HWE | N | Allele | P* | Genotype | MAF | P* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| T | C | T/T | T/C | C/C | |||||||
| F302 | SZ | 0.4533 | 944 | 975 | 913 | 246 | 483 | 215 | 48.4% | ||
| rs6425744 | CT | 0.1292 | 949 | 965 | 933 | 0.6259 | 257 | 451 | 241 | 49.2% | 0.2468 |
| Our SNP ID and rs# | HWE | N | Allele | P* | Genotype | MAF | P* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| T | C | T/T | T/C | C/C | |||||||
| F303 | SZ | 0.5241 | 944 | 1064 | 824 | 295 | 474 | 175 | 43.6% | ||
| rs10914367 | CT | 0.5100 | 948 | 1079 | 817 | 0.7429 | 312 | 455 | 181 | 43.1% | 0.6223 |
| Our SNP ID and rs# | HWE | N | Allele | P* | Genotype | MAF | P* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | G | A/A | A/G | G/G | |||||||
| F304 | SZ | 0.1950 | 942 | 1595 | 289 | 670 | 255 | 17 | 15.3% | ||
| rs11436 | CT | 0.0321 | 949 | 1583 | 315 | 0.3073 | 651 | 281 | 17 | 16.6% | 0.4655 |
| Our SNP ID and rs# | HWE | N | Allele | P* | Genotype | MAF | P* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | C | A/A | A/C | C/C | |||||||
| F305 | SZ | 0.3839 | 943 | 262 | 1624 | 15 | 232 | 696 | 13.9% | ||
| rs3766293 | CT | 0.7071 | 950 | 224 | 1676 | 0.0580 | 12 | 200 | 738 | 11.8% | 0.1390 |
| Our SNP ID and rs# | HWE | N | Allele | P* | Genotype | MAF | P* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | G | A/A | A/G | G/G | |||||||
| F306 | SZ | 0.9252 | 943 | 279 | 1607 | 21 | 237 | 685 | 14.8% | ||
| rs6663779 | CT | 0.3626 | 948 | 285 | 1611 | 0.8552 | 25 | 235 | 688 | 15.0% | 0.8512 |
| Our SNP ID and rs# | HWE | N | Allele | P* | Genotype | MAF | P* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | G | A/A | A/G | G/G | |||||||
| F307 | SZ | 0.9483 | 943 | 541 | 1345 | 78 | 385 | 480 | 28.7% | ||
| rs3795432 | CT | 0.9833 | 947 | 508 | 1386 | 0.2038 | 68 | 372 | 507 | 26.8% | 0.4391 |
| Our SNP ID and rs# | HWE | N | Allele | P* | Genotype | MAF | P* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| G | C | G/G | G/C | C/C | |||||||
| F308 | SZ | 0.5077 | 943 | 824 | 1062 | 175 | 474 | 294 | 43.7% | ||
| rs7532813 | CT | 0.5005 | 947 | 814 | 1080 | 0.6697 | 180 | 454 | 313 | 43.0% | 0.5818 |
- SZ, schizophrenia; CT, control; HWE, Hardy–Weinberg equilibrium; MAF, minor allele frequency.
- * Evaluated by Fisher's exact test.
| Our SNP ID and rs# | HWE | N | Allele | P* | Genotype | MAF | P* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| T | C | T/T | T/C | C/C | |||||||
| F301 | BP | 0.5503 | 860 | 572 | 1148 | 99 | 374 | 387 | 33.3% | ||
| rs12562824 | CT | 0.9101 | 890 | 642 | 1138 | 0.0819 | 115 | 412 | 363 | 36.1% | 0.1922 |
| Our SNP ID and rs# | HWE | N | Allele | P* | Genotype | MAF | P* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| T | C | T/T | T/C | C/C | |||||||
| F302 | BP | 0.4738 | 861 | 865 | 857 | 212 | 441 | 208 | 49.8% | ||
| rs6425744 | CT | 0.7450 | 893 | 859 | 927 | 0.2114 | 209 | 441 | 243 | 51.9% | 0.3406 |
| Our SNP ID and rs# | HWE | N | Allele | P* | Genotype | MAF | P* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| T | C | T/T | T/C | C/C | |||||||
| F303 | BP | 0.5951 | 861 | 961 | 761 | 272 | 417 | 172 | 44.2% | ||
| rs10914367 | CT | 0.9812 | 895 | 978 | 812 | 0.4973 | 267 | 444 | 184 | 45.4% | 0.7251 |
| Our SNP ID and rs# | HWE | N | Allele | P* | Genotype | MAF | P* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | G | A/A | A/G | G/G | |||||||
| F304 | BP | 0.3667 | 862 | 1432 | 292 | 591 | 250 | 21 | 16.9% | ||
| rs11436 | CT | 0.3966 | 894 | 1492 | 296 | 0.7863 | 619 | 254 | 21 | 16.6% | 0.9539 |
| Our SNP ID and rs# | HWE | N | Allele | P* | Genotype | MAF | P* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | C | A/A | A/C | C/C | |||||||
| F305 | BP | 0.0268 | 863 | 231 | 1495 | 23 | 185 | 655 | 13.4% | ||
| rs3766293 | CT | 0.5909 | 893 | 267 | 1519 | 0.1916 | 22 | 223 | 648 | 14.9% | 0.2176 |
| Our SNP ID and rs# | HWE | N | Allele | P* | Genotype | MAF | P* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | G | A/A | A/G | G/G | |||||||
| F306 | BP | 0.5076 | 862 | 253 | 1471 | 21 | 211 | 630 | 14.7% | ||
| rs6663779 | CT | 0.5767 | 893 | 260 | 1526 | 0.9239 | 21 | 218 | 654 | 14.6% | 1.0000 |
| Our SNP ID and rs# | HWE | N | Allele | P* | Genotype | MAF | P* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | G | A/A | A/G | G/G | |||||||
| F307 | BP | 0.1356 | 863 | 485 | 1241 | 77 | 331 | 455 | 28.1% | ||
| rs3795432 | CT | 0.7422 | 893 | 528 | 1258 | 0.3517 | 76 | 376 | 441 | 29.6% | 0.2734 |
| Our SNP ID and rs# | HWE | N | Allele | P* | Genotype | MAF | P* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| G | C | G/G | G/C | C/C | |||||||
| F308 | BP | 0.6382 | 861 | 762 | 960 | 172 | 418 | 271 | 44.3% | ||
| rs7532813 | CT | 0.9270 | 893 | 810 | 976 | 0.5189 | 183 | 444 | 266 | 45.4% | 0.7491 |
- BP, bipolar disorder; CT, control; HWE, Hardy–Weinberg equilibrium; MAF, minor allele frequency.
- * Evaluated by Fisher's exact test.
CNP of FABP3
Because CNP is frequently reported to be in LD with neighboring SNPs [Hinds et al., 2006], we selected 51 subjects who had different combinations of homozygous genotypes at F301 to F308 (i.e., all the SNP sites examined in the current study), to search for its existence (Table SII). However, none of them showed duplications or deletions of the FABP3 genomic region, suggesting that if present, this CNP is rare in the Japanese population.
DISCUSSION
PUFAs are integral components of membrane phospholipids and they are found abundantly in the brain. PUFAs are thought to be involved in multiple functions including cognition and emotion [Antypa et al., 2008]. Because PUFAs are insoluble in the intracellular matrix, specific transporters are required to deliver PUFAs to appropriate organelles. FABPs are believed to play crucial roles as their cellular shuttles.
In this study, we analyzed the three FABP genes expressed in the brain and detected association signals between FABP7 and bipolar disorder. A total of three SNPs (F704, F705, and F709) displayed allelic and genotypic associations with disease, although they were nominal. LD blocks 2 and 3 showed associations even after a gene-wide correction for multiple testing. Of the three SNPs, F05 is located in the associated LD block 2, but the other 2 SNPs were not in the associated LD blocks. This may be due to the differences in methods used to define tagging SNPs (r2) and LD blocks (D′) [Gabriel et al., 2002]. The three SNPs are in substantial LD to each other, especially in terms of D′ (Table SIII). For instance, the SNP F709 did not constitute a haplotype block under Gabriel's model [Gabriel et al., 2002] (Fig. 1). Since the extent of the haplotype block may delimit the range of a functional variant position, we reconstructed haplotype blocks using the solid spine model (D′ > 0.8). Under this model, the marker F709 was located within a block consisting of SNPs F707, F708, F709, F710, and F11, and the haplotype G–T–G–G–G was significantly over-represented in the bipolar disorder group (frequency = 0.36) compared to the control group (frequency = 0.32) [P = 0.014, OR (95% CI) = 1.19 (1.04–1.38)].
We also tested for an association between SNP F706 and schizophrenia, using the current expanded panel (the previously used sample set consisting of 570 schizophrenics and 570 controls). The results were: allelic P = 0.2352 and genotypic P = 0.2690, thus failing to replicate the prior finding. Because the minor allele frequency of this SNP is low [2.4% in schizophrenia and 3.1% in controls in the current panel; 1.7% in schizophrenia and 3.1% in controls in the previous panel] and the crystallographic analysis points to a probable functional alteration by this SNP [Watanabe et al., 2007], analysis of a much larger sample will be needed to draw a definite conclusion. In any case, further studies are needed to confirm the true causative SNPs and/or combination of SNPs in schizophrenia and bipolar disorder.
In our previous study, we demonstrated schizophrenia-related phenotypes in Fabp7 knockout mice, for example, reduced PPI and enhanced responses to repeated administration of MK-801 [Watanabe et al., 2007]. Based on these results, we are now examining emotion-related behavior in the gene-deficient mice. The results so far indicate elevated locomotor activity and enhanced anxiety traits in the knockout mice [unpublished data]. Therefore, although the human genetic data is modest, it may be possible that FABP7 does have some role in the development of schizophrenia and bipolar disorder. It is interesting to note that Fabp7 shows abundant expression in neural progenitor cells during early developmental stages and augments neurogenesis [Arai et al., 2005; Watanabe et al., 2007; Owada, 2008]. The potential links between neurogenesis and mood disorder [see Eisch et al., 2008 for review] and schizophrenia [Reif et al., 2006] have been reported. Therefore if altered neurogenesis is a contributory mechanism to the pathogenesis of schizophrenia and bipolar disorder, FABP7 may be a strong causative gene. Regarding the relationship between PUFAs and mood disorders, another line of evidence is also notable: administration of three mood stabilizers (lithium, valproate, and carbamazepine) at therapeutically relevant doses, selectively target the brain arachidonic acid cascade, and decrease turnover of arachidonic acid but not of docosahexaenoic acid in rat brain [Rao et al., 2008].
The structure of each FABP gene has been conserved among all members of the family; they consist of four exons separated by three introns [Veerkamp and Zimmerman, 2001]. One of the impediments in genetic studies of FABP genes is the relatively small size of FABP7 (=4.57 kb), FABP5 (=4.22 kb), and FABP3 (=7.82 kb). We could not obtain suitable SNPs for FABP5, even though we expanded the region of our search for polymorphisms to 10 kb-upstream and 10 kb-downstream from the first exon and last exon (re-sequencing analysis plus database search). Functionally, FABP5 shares similarities with FABP7, in terms of their ontogenic expression patterns [Owada, 2008] and roles in neurogenesis [unpublished data]. In contrast, the expression of Fabp3 in the brain increases slowly in postnatal stages, reaching a plateau in adulthood [Owada, 2008]. Interestingly in relation to psychiatric illnesses, Fabp3 co-localizes with dopamine receptor positive cells, and it interacts with the dopamine receptor D2L, and regulates the distribution of the D2L between the membrane and perinuclear cytoplasm [Takeuchi and Fukunaga, 2003].
Expression of each FABP gene is spatio-temporally regulated very tightly, using multiple regulatory elements in addition to the core promoter [Haunerland and Spener, 2004]. However, none of these regulatory genomic elements have been identified. For a more comprehensive evaluation of the genetic contribution of FABP genes to schizophrenia and bipolar disorder, future studies are needed to clarify such genomic elements and assess the roles of polymorphisms found in those regions.
Acknowledgements
The authors would like to acknowledge all the subjects who participated in this study. This work was supported by RIKEN BSI Funds, CREST funds from the Japan Science and Technology Agency, and grants from MEXT of Japan. The authors report no involvement, financial or otherwise, that might potentially bias this work.




