GABRR1 and GABRR2, encoding the GABA-A receptor subunits ρ1 and ρ2, are associated with alcohol dependence†
How to Cite this Article: Xuei X, Flury-Wetherill L, Dick D, Goate A, Tischfield J, Nurnberger J, Jr, Schuckit M, Kramer J, Kuperman S, Hesselbrock V, Porjesz B, Foroud T, Edenberg HJ. 2009. GABRR1 and GABRR2, Encoding the GABA-A Receptor Subunits ρ1 and ρ2, Are Associated With Alcohol Dependence. Am J Med Genet Part B 153B:418–427.
Abstract
The genes encoding several GABA-A receptor subunits, including GABRA2, have been associated with alcoholism, suggesting that variations in gaba signaling contribute to risk. Therefore, as part of a comprehensive evaluation of the GABA receptor genes, we evaluated the potential association of GABRR1 and GABRR2, which encode the ρ1 and ρ2 subunits of the pentameric GABA-A/GABA-C receptors. GABRR1 and GABRR2 lie in a head to tail orientation spanning 137 kb on chromosome 6q14-16. We genotyped 73 single nucleotide polymorphisms (SNPs), covering both genes and extending 31 kb upstream of GABRR2 and 95 kb downstream of GABRR1, in a sample of 1923 European Americans from 219 multiplex alcohol-dependent families. Family-based association analyses demonstrated that SNPs in both GABRR1 and GABRR2 were significantly associated with alcohol dependence. Among the associated SNPs was rs282129, a coding SNP (Met430Thr) in GABRR2. Secondary analysis using a median split for age of onset suggests that the association is strongest when the analysis is focused upon those with earlier onset of alcohol dependence. Haplotypes in each gene were significantly overtransmitted to family members who did not meet criteria for alcohol dependence (P < 0.04), and a haplotype in GABRR2 was significantly overtransmitted to family members who met a broader definition of alcoholism (P = 0.002) as well as DSM-IV dependence (P = 0.04). © 2009 Wiley-Liss, Inc.
INTRODUCTION
Alcoholism is a complex genetic disorder, affected by multiple genes, environmental and social influence, and interactions between genes and environmental factors [Cloninger et al., 1981; Kendler et al., 1994; Heath et al., 1997; Bierut et al., 1999]. To identify genes contributing to the risk for alcoholism, the Collaborative Study on the Genetics of Alcoholism (COGA) has utilized both linkage and association approaches, using carefully ascertained and rigorously evaluated individuals in families with multiple alcoholics [Begleiter et al., 1995; Edenberg, 2002; Foroud et al., 2000]. With this approach, COGA has provided evidence that a number of specific genes are associated with the risk for alcoholism, including GABRA2, CHRM2, hTAS2R16, GABRG3, OPRK1, PDYN, ADH4, NFKB1, TACR3, NPY2R, and ACN9 [Dick et al., 2004; Edenberg et al., 2004, 2006, 2008; Wang et al., 2004; Hinrichs et al., 2006; Xuei et al., 2006; Dick et al., 2008; Foroud et al., 2008; Wetherill et al., 2008].
GABA-A receptors are ligand-gated anion channels formed by pentameric complexes including α, β, γ, δ, ε, π, and ρ subunits, and mediate fast synaptic inhibition in response to GABA [Cutting et al., 1991; Enoch, 2008]. GABA binds to an extracellular ligand-binding domain, while four transmembrane domains form the ion channel for chloride transportation into cells. The effects of drugs and alcohol on GABA neurotransmission have been extensively studied, as has their role in alcohol dependence [Barnard et al., 1998; Koob, 2006; Lobo and Harris, 2008; Vengeliene et al., 2008]. The GABA-ρ1 receptor subunit was originally cloned based upon homology to the GABA-A receptor subunits, and when expressed encodes a GABA-responsive chloride channel that binds mucimol [Cutting et al., 1991]. The closely related ρ2 subunit was cloned by homology to ρ1 [Cutting et al., 1992]. Sequence similarity makes them part of the GABA-A family of receptors, and they form ligand-gated chloride channels, but insensitivity of the homopentamers to baclofen and bicuculline as well as benzodiazepines lead them to also be classified as GABA-C receptors [Shimada et al., 1992; Kusama et al., 1993; Bormann, 2000; Zhang et al., 2001]. GABA-C receptors have a higher sensitivity to GABA and do not desensitize [Enz, 2001]. In the superior colliculus they appear to be involved in long-term potentiation [Enz, 2001; Schmidt et al., 2001].
Several GABA-A receptor genes are associated with alcoholism. The association of GABRA2 with alcoholism [Edenberg et al., 2004] has been replicated in case–control studies of Germans [Fehr et al., 2006; Soyka et al., 2008], Russians [Lappalainen et al., 2005], European-Americans [Covault et al., 2004], Plain Indians [Enoch et al., 2006], and a general Australian population [Lind et al., 2008]. The association was with single nucleotide polymorphisms (SNPs) in a region of linkage disequilibrium (LD) extending from intron 3 past the 3′ end of the GABRA2 gene [Covault et al., 2004; Edenberg et al., 2004] and may extend to the 5′ end of the adjacent GABRG1 gene, which is in LD with SNPs in GABRA2 [Covault et al., 2008]. In the COGA sample, the association with GABRA2 was greatest among those alcohol-dependent individuals with comorbid dependence on illicit drugs [Agrawal et al., 2006]; this subgroup is characterized by greater severity of alcohol problems [Dick et al., 2007]. SNPs in GABRG3 on chromosome 15 [Dick et al., 2004] and GABRA1 on chromosome 5 [Dick et al., 2006] have also been associated with alcoholism. The evidence for associations of several GABA receptors with alcoholism, along with the known effects of alcohol on GABA neurotransmission, raises the question of whether variations in additional GABA-A receptors, including ρ receptors, might also affect risk for alcoholism.
There is evidence for linkage of the power in the gamma band (29–45 Hz) of the resting electroencephalograph (EEG) over the frontal and central scalp with markers on chromosome 6q14-16 [Xuei et al., 2008]. GABRR1 and GABRR2, encoding the GABA receptor subunits ρ1 and ρ2, lie together in a 137 kb region that falls within this linkage peak. Although the ρ1 and ρ2 subunits were originally found in the bipolar neurons in the retina [Cutting et al., 1991; Polenzani et al., 1991], they are widely expressed in the brain (including cortex, thalamus, pituitary gland, cerebellum, and hippocampus) and in the spinal cord [Cutting et al., 1991; Zheng et al., 2003; Milligan et al., 2004; Lopez-Chavez et al., 2005; Alakuijala et al., 2005a,b; Harvey et al., 2006]. Homomeric ρ1 receptors are inhibited by ethanol at a low concentration (400 nM) of GABA [Mihic and Harris, 1996]. Two amino acids in the GABA-A receptor transmembrane domains 2 (Ser270) and 3 (Ala291) are critical for allosteric modulation of the receptors by alcohols and volatile anesthetics [Mihic et al., 1997]. Heteromeric complexes of ρ and α1 subunits were found in mouse cerebellar Purkinje cells, where ρ subunits contribute to functional ionotropic receptors mediating a component of phasic inhibitory GABAergic transmission [Harvey et al., 2006].
Therefore, we examined whether GABRR1 and GABRR2 might be associated with alcoholism. We genotyped 73 SNPs, covering GABRR1, GABRR2, and their flanking regions, in a sample of European Americans from families in which at least three first-degree relatives were alcohol dependent. We report here evidence that SNPs in both GABRR1 and GABRR2 were significantly associated with alcohol dependence.
MATERIALS AND METHODS
Subjects
Subjects were collected at six centers in the United States: Indiana University, State University of New York Health Science Center, University of Connecticut, University of Iowa, University of California/San Diego, and Washington University, St. Louis. The Institutional Review Boards of all participating institutions approved the study. Probands were identified through alcohol treatment programs. After providing informed consent, probands and their relatives were administered a validated poly-diagnostic instrument, the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA) interview [Bucholz et al., 1994; Hesselbrock et al., 1999]. For ascertainment, alcoholism was defined as meeting criteria for both DSM-IIIR alcohol dependence [American Psychiatric Association, 1987] and Feighner definite alcoholism [Feighner et al., 1972] (this combination is called COGA criteria). Details of the ascertainment and assessment have previously been published [Begleiter et al., 1995; Reich et al., 1998; Foroud et al., 2000] and are available in detail at zork.wustl.edu/niaaa/coga_instruments/resources.html. Families in which at least three first-degree relatives were alcohol dependent participated in the genetic part of the COGA study, as described in more detail by Foroud et al. 2000. A sample of 1923 European American individuals from 219 alcoholic families was used in this study.
SNP Genotyping
Because HapMap data indicated that flanking genes were in LD with the GABRR1–GABRR2 gene cluster, we extended the genotyping into the flanking regions. Seventy-three SNPs in a 262 kb region, including GABRR1 and GABRR2, UBE2J1 (ubiquitin-conjugating enzyme E2, J1) upstream of GABRR2, and PM20D2 (peptidase M20 domain containing 2), SRrp35 (serine–arginine repressor protein), and PNRC1 (proline-rich nuclear receptor coactivator 1) downstream of GABRR1 (Fig. 1), were selected from public databases (dbSNP and HapMap). Preference was given to SNPs having minor allele frequencies greater than 5%. Four synonymous SNPs in the coding regions of GABRR1 and GABRR2 (rs34218666, rs35608866, rs34617047, and rs35301635) were genotyped despite their reported low minor allele frequencies; they proved monomorphic in our sample and are not included in the tables or analyses. SNP positions were obtained from NCBI reference human genome, build 36.3.
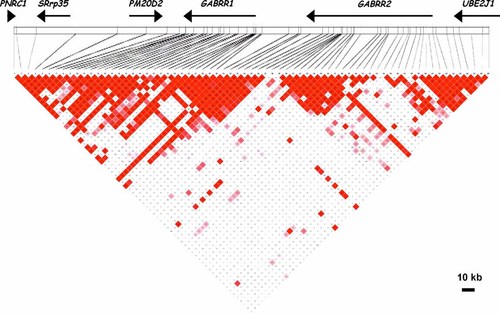
Locations of genes, SNPs, and linkage disequilibrium (D′) among the SNPs. Sixty nine SNPs (MAF > 0.05) are included in the plot of D′. The location of each gene is represented proportionally across the top of the figure, with the direction of transcription represented by an arrow. The size of the gene is indicated at lower right side.
Most assays were designed for the Sequenom MassArray system (Sequenom, San Diego, CA) using MassArray Assay Design Software. The assays were done using the hME or iPLEX assay format (Sequenom); in both cases, alleles were discriminated by mass spectrometry. Assays were tested on two groups of 40 unrelated individuals from the Coriell European-American and African-American samples. SNPs that were not in Hardy–Weinberg equilibrium in both populations were not genotyped in the sample. Some SNPs were genotyped using the Illumina technology on a BeadLab station with GoldenGate chemistry, as described previously [Dick et al., 2008].
All SNPs were tested for Mendelian inheritance using the program PEDCHECK [O'Connell and Weeks, 1998]. Marker allele frequencies and heterozygosities were computed using the program USERM13 [Boehnke, 1991]. Markers were tested for Hardy–Weinberg equilibrium in independent individuals from the study sample using Haploview [Barrett et al., 2005] and were omitted from analysis if they deviated significantly (P < 0.001) from Hardy–Weinberg equilibrium.
Statistical Analyses
LD among genotyped SNPs was evaluated using HAPLOVIEW [Barrett et al., 2005]. Coverage of the gene was additionally examined using Tagger [de Bakker et al., 2005] to determine the correlation between genotyped SNPs and all known SNPs in the HapMap I dataset.
Family-based association analyses were performed using the Pedigree Disequilibrium Test (PDT) [Martin et al., 2001] as implemented in the program UNPHASED (version 2.404) [Dudbridge, 2003]. The PDT uses data from all available trios in a family, as well as discordant sibships. The PDTsum statistic, which gives greater weight to families with a larger number of informative trios and discordant sibships, was utilized as the primary statistic. Haplotypes from phase-certain individuals were used to further explore significant associations across the GABRR1–GABRR2 gene cluster. Association analysis using the haplotypes was performed using UNPHASED and the PDTsum statistic.
For our primary phenotype, we employed the DSM-IV criteria for alcohol dependence [American Psychiatric Association, 1994], which could be derived from the SSAGA data; we used the broader COGA definition as a secondary phenotype. The vast majority of individuals who met DSM-IV criteria also met COGA criteria; 35 individuals who were positive for DSM-IV but negative for COGA criteria and 22 individuals who were positive for COGA criteria but negative for DSM-IV were coded as unknown and omitted from analysis (Table I).
| Phenotypes | # Affected (%) | # Unaffected (%) | Unknowna (%) |
|---|---|---|---|
| DSM-IV | 718 (37.3%) | 1025 (53.3%) | 180 (9.4%) |
| COGA | 862 (44.8%) | 881 (45.8%) | 180 (9.4%) |
| Earlyb | 399 (20.7%) | 1025 (53.3%) | 499 (26.0%) |
- a There are 123 individuals without a completed SSAGA diagnostic interview, and 57 individuals recoded as unknown due to contradicting alcohol dependence classification.
- b Subjects coded as early met the DSM-IV criteria for alcohol dependence at age 21 or younger.
We performed secondary analyses exploring the hypothesis that genetic effects are stronger in those alcohol-dependent individuals with an earlier age of onset. Association analysis was performed defining as affected only those individuals meeting DSM-IV criteria for alcohol dependence by age 21 (n = 399; Table I). As in the above analyses of alcohol dependence, 1,025 family members were classified as unaffected. Others, including individuals meeting DSM-IV alcohol dependence criteria at age 22 or older, were considered unknown.
RESULTS
GABRR1 and GABRR2 are in a head-to-tail orientation spanning 137 kb on chromosome 6q (Fig. 1). Based on the LD pattern, we analyzed 69 SNPs in the region extending 31 kb upstream of GABRR2 to the adjacent gene UBE2J1, and 95 kb downstream of GABRR1; this 263 kb region included three additional genes, PM20D2, SRrp35, and PNRC1 (Fig. 1). The mean minor allele frequency (MAF) was 0.29, with a standard deviation ±0.11 and a range from 0.06 to 0.50 (Table II). The LD pattern in our sample is similar to that in the HapMap CEU (CEPH European) database.
| Gene | SNP number | SNP_ID | Chromosome positiona | SNP locationb | MAFc | Minor nucleotided | DSM-IVe | Early onsete | COGAe |
|---|---|---|---|---|---|---|---|---|---|
| PNRC1 | 1 | rs1130809 | 89,850,613 | Exon 2, Thr321 | 0.18 | C | 0.70 | 0.18f | 0.77 |
| 2 | rs11961455 | 89,852,007 | Downstream | 0.42 | C | 0.66 | 0.23f | 0.39 | |
| SRrp35 | 3 | rs6938490 | 89,862,949 | 3′ UTR | 0.29 | A | 0.90 | 0.47 | 0.33 |
| 4 | rs423516 | 89,884,774 | Promoter | 0.29 | T | 0.57 | 0.15 | 0.72 | |
| PM20D2 | 5 | rs4707518 | 89,907,725 | Upstream | 0.29 | C | 0.69 | 0.22 | 0.98 |
| 6 | rs1929635 | 89,923,709 | Intron 4 | 0.28 | G | 0.49 | 0.13 | 0.73 | |
| GABRR1 | 7 | rs1331100 | 89,935,180 | Downstream | 0.29 | G | 0.72 | 0.18 | 0.81 |
| 8 | rs416115 | 89,941,956 | Downstream | 0.30 | C | 0.17 | 0.014f | 0.08f | |
| 9 | rs3734201 | 89,945,162 | 3′ UTR | 0.42 | C | 0.58 | 0.04 | 0.55 | |
| 10 | rs1796743 | 89,945,463 | Exon 10, Ala389 | 0.35 | T | 0.40 | 0.12f | 0.27 | |
| 11 | rs407221 | 89,947,094 | Intron 8 | 0.27 | C | 0.21 | 0.23 | 0.37 | |
| 12 | rs368873 | 89,949,881 | Intron 7 | 0.29 | G | 0.43 | 0.03f | 0.14f | |
| 13 | rs407206 | 89,951,687 | Intron 7 | 0.32 | A | 0.13 | 0.12 | 0.15 | |
| 14 | rs423463 | 89,955,427 | Intron 6 | 0.19 | A | 0.68 | 0.41f | 0.97 | |
| 15 | rs453503 | 89,957,318 | Intron 5 | 0.19 | T | 0.64 | 0.38f | 0.66 | |
| 16 | rs452667 | 89,960,039 | Intron 5 | 0.19 | T | 0.62 | 1.00 | 0.45 | |
| 17 | rs439912 | 89,963,734 | Intron 5 | 0.45 | C | 0.13 | 0.56 | 0.32 | |
| 18 | rs422751 | 89,964,592 | Exon 5, Asp140 | 0.16 | G | 0.97 | 0.86 | 0.8 | |
| 19 | rs11967322 | 89,966,483 | Intron 3 | 0.43 | T | 0.19 | 0.45 | 0.58 | |
| 20 | rs7741132 | 89,969,614 | Intron 2 | 0.33 | A | 0.14 | 0.06 | 0.13 | |
| 21 | rs7758893 | 89,973,360 | Intron 1 | 0.19 | C | 0.79 | 0.51f | 0.76 | |
| 22 | rs881293 | 89,974,522 | Intron 1 | 0.32 | C | 0.63 | 0.29f | 0.71 | |
| 23 | rs1029057 | 89,979,273 | Intron 1 | 0.28 | T | 1.00 | 0.87 | 0.44 | |
| 24 | rs6902106 | 89,982,459 | Intron 1 | 0.28 | C | 0.34 | 0.14f | 0.09f | |
| 25 | rs17504587 | 89,982,921 | Intron 1 | 0.19 | T | 0.04 | 0.03 | 0.01 | |
| 26 | rs1186902 | 89,983,681 | Exon 1, Arg21His | 0.28 | C | 0.81 | 0.97 | 0.36 | |
| 27 | rs12200969 | 89,983,685 | Exon 1, Val20Met | 0.34 | C | 0.20 | 0.09f | 0.11f | |
| 28 | rs1186903 | 89,984,290 | Promoter | 0.35 | A | 0.11f | 0.048f | 0.10f | |
| 29 | rs914479 | 89,986,927 | Upstream | 0.35 | C | 0.08f | 0.028f | 0.04f | |
| 30 | rs914478 | 89,987,638 | Upstream | 0.34 | C | 0.12f | 0.06f | 0.07f | |
| 31 | rs9342188 | 90,002,245 | Intergenic | 0.09 | C | 0.83 | 0.27f | 0.59 | |
| 32 | rs1321355 | 90,007,632 | Intergenic | 0.27 | G | 0.10f | 0.10f | 0.08f | |
| 33 | rs282135 | 90,008,949 | Intergenic | 0.28 | G | 0.74 | 0.93 | 0.24 | |
| 34 | rs6914006 | 90,014,480 | Intergenic | 0.44 | A | 0.46 | 0.47 | 0.73 | |
| GABRR2 | 35 | rs3734198 | 90,021,011 | Downstream | 0.43 | G | 0.62 | 0.84 | 0.92 |
| 36 | rs282129 | 90,024,217 | Exon 9, Met430Thr | 0.28 | A | 0.03f | 0.08f | 0.02f | |
| 37 | rs13211104 | 90,027,125 | Intron 8 | 0.30 | A | 0.03f | 0.07f | 0.03f | |
| 38 | rs9451191 | 90,029,831 | Intron 8 | 0.26 | A | 0.021 | 0.002 | 0.17 | |
| 39 | rs723041 | 90,030,782 | Intron 8 | 0.13 | T | 0.79 | 0.86 | 0.83 | |
| 40 | rs9294426 | 90,031,297 | Intron 7 | 0.29 | A | 0.55 | 0.38 | 1.00 | |
| 41 | rs12206367 | 90,031,345 | Intron 7 | 0.12 | A | 0.43 | 0.38 | 0.84 | |
| 42 | rs2273507 | 90,031,964 | Intron 7 | 0.20 | G | 0.70 | 0.55 | 0.52 | |
| 43 | rs2273508 | 90,034,567 | Intron 4 | 0.11 | T | 0.97 | 0.63 | 0.46 | |
| 44 | rs282121 | 90,036,802 | Intron 3 | 0.41 | T | 0.025 | 0.02 | 0.05 | |
| 45 | rs282117 | 90,038,132 | Exon 3, Val83 | 0.42 | T | 0.07 | 0.017 | 0.12 | |
| 46 | rs282115 | 90,038,431 | Intron 2 | 0.07 | A | 0.78 | 0.20f | 0.96 | |
| 47 | rs9451194 | 90,042,098 | Intron 2 | 0.45 | A | 0.91 | 0.53 | 0.65 | |
| 48 | rs9362632 | 90,051,417 | Intron 2 | 0.29 | G | 0.87 | 0.88 | 0.36 | |
| 49 | rs3798256 | 90,053,124 | Intron 2 | 0.45 | G | 0.22 | 0.06 | 0.36 | |
| 50 | rs964626 | 90,056,015 | Intron 2 | 0.21 | C | 0.13f | 0.16f | 0.13f | |
| 51 | rs9451196 | 90,057,273 | Intron 2 | 0.12 | T | 0.07f | 0.21f | 0.30f | |
| 52 | rs6454750 | 90,057,490 | Intron 2 | 0.45 | T | 1.00 | 0.65 | 0.80 | |
| 53 | rs2148174 | 90,062,644 | Intron 2 | 0.33 | T | 0.41 | 0.78 | 0.85 | |
| 54 | rs7742664 | 90,065,463 | Intron 2 | 0.22 | T | 0.20f | 0.03f | 0.25f | |
| 55 | rs2325202 | 90,068,229 | Intron 1 | 0.36 | G | 0.22 | 0.04f | 0.16 | |
| 56 | rs1570028 | 90,069,154 | Intron 1 | 0.49 | A | 0.34 | 0.07f | 0.08 | |
| 57 | rs6942204 | 90,072,687 | Intron 1 | 0.27 | C | 0.04 | 0.005 | 0.016 | |
| 58 | rs6454752 | 90,075,080 | Intron 1 | 0.23 | T | 0.43 | 0.21f | 0.07f | |
| 59 | rs9294432 | 90,076,686 | Intron 1 | 0.47 | T | 0.51 | 0.14f | 0.04f | |
| 60 | rs10944441 | 90,078,260 | Intron 1 | 0.18 | A | 0.08f | 0.012f | 0.005f | |
| 61 | rs7764923 | 90,079,640 | Intron 1 | 0.24 | T | 0.08f | 0.02f | 0.007f | |
| 62 | rs2236204 | 90,081,831 | Promoter | 0.19 | T | 0.09f | 0.015f | 0.006f | |
| 63 | rs9362636 | 90,083,516 | Promoter | 0.06 | G | 0.86 | 0.88 | 0.91 | |
| 64 | rs2064831 | 90,089,661 | Upstream | 0.18 | C | 0.09f | 0.01f | 0.009f | |
| UBE2J1 | 65 | rs13215418 | 90,093,230 | Downstream | 0.17 | T | 0.56 | 0.93 | 0.73 |
| 66 | rs10502 | 90,096,389 | Exon 8, Val229Leu | 0.40 | C | 0.34 | 0.11f | 0.07f | |
| 67 | rs12189673 | 90,103,123 | Intron 5 | 0.30 | C | 0.62 | 0.25 | 0.05 | |
| 68 | rs7760851 | 90,109,323 | Intron 2 | 0.50 | G | 0.40 | 0.48 | 0.009 | |
| 69 | rs7743444 | 90,112,800 | Intron 1 | 0.24 | T | 0.88 | 0.97 | 0.52 |
- a Chromosome positions are based on NCBI Human Genome Assembly v36.3.
- b PNRC1 and PM20D2 are transcribed in forward direction; SRrp35, GABRR1, GABRR2, and UBE2J1 are transcribed in the opposite direction.
- c Minor allele frequency in this sample of European Americans.
- d Nucleotides are shown on the human genome strand.
- e P-value of UNPHASED PDTsum statistic for association between the SNP and alcohol dependence. P ≤ 0.05 is in bold.
- f Minor allele is preferentially transmitted to affected individuals.
SNP coverage was further assessed using Tagger [de Bakker et al., 2005]; 61 of the SNPs were included in the HapMap database and could, therefore, be analyzed. In the CEU population, these 61 SNPs gave a mean r2 of 0.79 with a total of 250 HapMap SNPs in the region; 65% of the SNPs in the region were captured at r2 ≥ 0.8 and 84% at r2 ≥ 0.5. The eight additional SNPs we genotyped that were not included in the HapMap database would further increase the coverage of this region. Thus, the SNPs we genotyped provide very good coverage of the variation in the GABRR1–GABRR2 gene cluster and adjacent genes.
Six SNPs were significantly associated with alcohol dependence defined by DSM-IV criteria (P < 0.05) (Table II, Fig. 2); five of these were located in GABRR2 from intron 1 to exon 9 and 1 was in intron 1 of GABRR1. Interestingly, rs282129 (P = 0.03) is a nonsynonymous coding SNP that leads to either a methionine or a threonine at amino acid 430 of GABRR2; the allele overtransmitted to the affected individuals encodes methionine. Eight additional SNPs (including four in the promoter to intron 1 region of GABRR2 and 1 upstream of GABRR1) demonstrated suggestive association (0.05 < P ≤ 0.10).
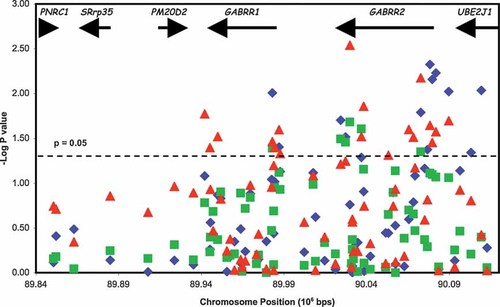
Association of GABRR1 and GABRR2 with alcohol dependence. The −log (P-value) of the PDTsum statistic for each SNP is plotted as a function of chromosomal position (106 bp). The solid squares represent alcohol dependence using DSM-IV criteria; the solid triangles represent early onset alcohol dependence; the solid diamonds represent alcohol dependence using COGA criteria. Dashed line indicates P-value = 0.05. The gene locations are shown across the top.
We have previously found that the associations of GABRA2, CHRM2, and NFKB1 with alcoholism were strongest in the half of the sample that included the most severely affected individuals, whose onset of alcohol dependence was at age 21 or younger [Agrawal et al., 2006; Dick et al., 2007; Edenberg et al., 2008]. Therefore, we performed additional analysis using a median split for age of onset of alcohol dependence, defining affected individuals as those who met DSM-IV criteria for alcohol dependence at age 21 or earlier (n = 399). Despite the reduction in power due to the substantially lower number of affected individuals, greater evidence for association was observed, with 16 significantly associated SNPs (P ≤ 0.05) in the GABRR1 and GABRR2 gene cluster, including a synonymous SNP (Val83; rs282117) in GABRR2, and another 8 suggestive (0.05 < P ≤ 0.10; Table II, Fig. 2).
Secondary analyses using the broader COGA criteria for alcohol dependence provided evidence for association; a total of 13 SNPs, including 8 concentrated in the region from intron 1 of GABRR2 upstream to a part of UBE2J1 that was in LD with the 5′ region of GABRR2, and 2 in the region from just upstream to intron 1 of GABRR1 (Table II, Fig. 2).
The pattern of LD was examined in each of the regions having SNPs associated with at least one of the three phenotypes (DSM-IV alcohol dependence, early onset DSM-IV alcohol dependence, and COGA alcohol dependence). While there was extensive LD as defined by D′ (Fig. 1), differences in allele frequencies mean that the correlation of the SNPs (r2) was much more restricted (Fig. 3), such that different groups of SNPs were correlated with each other but much less with other groups. Considering the LD pattern, it appears that there are at least six independent regions of association. Several groups of SNPs in LD with each other show similar patterns of association. At the 3′ end of GABRR1, SNPs 8, 10, and 12 are in high LD with each other (Fig. 3, inset) and two provide evidence of association with early onset, while the third, less correlated SNP, approaches significance; SNP 9, in that cluster, is also significant. At the 5′ end of GABRR1, three SNPs (25, 28, 29) are significantly associated with early onset (and several others are nearly significant), with some evidence for association with the COGA definition of dependence (Table II, Fig. 3). Three SNPs (36–38) at the 3′ end of GABRR2 (which is just upstream of the 5′ end of GABRR1, Fig. 3) show evidence for association with all three phenotypes, particularly DSM-IV dependence. Two of these, including the cSNP at position 430, are in LD with the group of SNPs at the 5′ end of GABRR1 (28–32; Fig. 3, inset). Thus these two groups of SNPs may represent a single finding. SNP 38 is not in LD with other SNPs, and is most significant for the early onset phenotype.
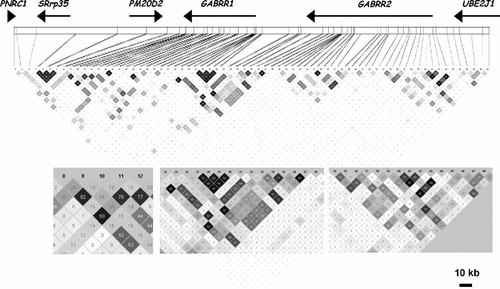
Linkage disequilibrium (r2) among the SNPs. The correlation among SNPs (r2) across the region is shown in the large figure; insets magnify three regions discussed in the text.
One SNP from each of these latter two groups was selected (rs914479 and rs282129; r2 = 0.42) and used in haplotype analysis to test whether a high risk haplotype could be identified within the region spanned by SNPs 27–37. The global test of significance was significant for all three alcohol dependence phenotypes: DSM-IV (P = 0.016), COGA (P = 0.014), and early onset (P = 0.030) (Table IIIa). For all three phenotypes, a common haplotype having a frequency of 61% (T-G) was associated with the absence of alcohol dependence P < 0.008 for DSM-IV; P < 0.007 for COGA; P = 0.019 for early onset. The complementary haplotype C-A, consisting of the minor alleles of both SNPs, was associated with COGA alcohol dependence (P = 0.02) and showed a trend toward significant association with DSM-IV (P = 0.11); the low frequency of this haplotype gives it lower power.
| rs914479 nucleotidea | rs282129 nucleotidea | DSM-IVc | Early onsetc | COGAc |
|---|---|---|---|---|
| T | G | 0.008 | 0.019 | 0.007 |
| T | Ab | 0.08 | 0.07 | 0.43 |
| Cb | G | 0.33 | 0.12 | 0.34 |
| Cb | Ab | 0.11 | 0.35 | 0.023 |
| Global test | 0.016 | 0.03 | 0.014 | |
- a Nucleotides are shown on the human genome strand.
- b Minor allele.
- c P-value of UNPHASED PDTsum statistic for association between the haplotype and alcohol dependence. P ≤ 0.05 is in bold.
There are two other clusters of SNPs in GABRR2 (SNPs 44–49 and 54–64) with high LD within each group (r2 > 0.54 and r2 > 0.64 respectively) but little LD between the two groups. There is consistency in the phenotypes associated with the significant SNPs and with the pattern of LD. One SNP from each group was selected for haplotype analysis (rs282121 and rs10944441; r2 = 0.06). The global test of association was significant for the broader COGA phenotype (P = 0.005) and for early onset (P = 0.05) alcohol dependence, and marginally significant for DSM-IV alcohol dependence (P = 0.07) (Table IIIb). Haplotype T-G (frequency 38%), consisting of the minor allele of rs282121 and the major allele of rs10944441, was overtransmitted to individuals not meeting any of the three alcohol dependence criteria and its complement, C-A (frequency 12%), was overtransmitted to the individuals meeting COGA (P = 0.002) and DSM-IV (P = 0.044) criteria for alcohol dependence, but not related to early onset.
| rs282121 nucleotidea | rs1094441 nucleotidea | DSM-IVc | Early onsetc | COGAc |
|---|---|---|---|---|
| Tb | Ab | 0.84 | 0.30 | 0.24 |
| Tb | G | 0.035 | 0.014 | 0.012 |
| C | Ab | 0.044 | 0.17 | 0.002 |
| C | G | 0.37 | 0.25 | 0.67 |
| Global test | 0.07 | 0.05 | 0.005 | |
- a Nucleotides are shown on the human genome strand.
- b Minor allele.
- c P-value of UNPHASED PDTsum statistic for association between the haplotype and alcohol dependence. P ≤ 0.05 is in bold.
DISCUSSION
This is the first study reporting evidence of association between GABRR1 and GABRR2 and alcohol dependence. Our primary analysis, using DSM-IV criteria as the phenotype, gave evidence of association primarily in GABRR2 (five significant and six suggestive SNPs; Fig. 1, Table II). Among the associated SNPs, rs282129 is a nonsynonymous coding SNP in exon 9 of GABRR2; the major allele (G) encodes Thr430 in transmembrane domain 4, which forms part of the ion channel, and the minor allele (A) encodes Met430 (nucleotides are shown on the human genome strand). Met is overtransmitted to alcoholic individuals, suggesting that the protein with Met430 might differ in function in a way that could contribute to the development of alcohol dependence. However, this cSNP is in a larger region of LD extending to the upstream region of GABRR1, thus it could be that other regulatory variations in the region are responsible for the association.
Because early onset of alcoholism often reflects greater severity, including a higher risk for antisocial personality disorder and conduct disorder [Dick et al., 2007], and some of our earlier findings were stronger with this sub-phenotype [Agrawal et al., 2006; Dick et al., 2007; Edenberg et al., 2008], we examined whether the findings were primarily due to the cases with early onset. This was, in fact, found: 18 SNPs spread across the 2-gene cluster, including a synonymous coding SNP (rs282117; Val83) in GABRR2, were significantly associated with early onset alcohol dependence. The increase in evidence for association despite the reduction by nearly half in the number of individuals deemed affected in this analysis suggests that the association with alcohol dependence is particularly strong in individuals who developed alcoholism at age 21 or younger.
Secondary analysis using a broader definition of alcoholism that includes additional individuals (COGA definition; Table I) shows more SNPs significantly associated, particularly in the 5′ portion of GABRR2. The associated SNPs within each portion of GABRR1 and GABRR2 are in LD with each other (Fig. 3). Both regions of association cover the 5′ promoter to the intron 1 region of each gene, suggesting that they may affect the regulation of these genes. Several SNPs in the two associated regions (SNPs 25–27 and 36, and SNPs 60–62) are located in highly conserved regions, suggesting potential functional roles in gene expression. The two intronic SNPs in UBE2J1 (SNPs 67 and 68) are in LD with the upstream region of GABRR2 (Fig. 3), and may also affect expression of GABRR2.
The pattern of LD across these genes suggests that there are at least six different regions of association (Fig. 3), and that there may be several different susceptibility haplotypes in this region. Between the 5′ region of GABRR1 and the 3′ end of GABRR2, we detected a haplotype (rs914479[T]–rs282129[G]) that appears to confer protection against all three phenotypes of alcohol dependence (Table III). This region includes the coding SNP rs282129. A haplotype within the coding and proximal 5′ portion of GABRR2 (rs282121[T]–rs1094441[G]) also appears to confer protection from alcohol dependence (Table III). We detected strong evidence of association of the complementary haplotype rs282121[C]–rs1094441[A] to COGA criteria for alcohol dependence (P = 0.002), less evidence of association with the narrower DSM-IV criteria (P = 0.044) but no evidence of association with the early onset subset of our sample; this pattern suggests that the C-A haplotype confers risk for alcoholism but not particularly for the more severe manifestations.
We analyzed one primary phenotype, DSM-IV alcohol dependence, and have performed secondary analyses using two phenotypes that included as affected only those meeting criteria for early onset DSM-IV alcohol dependence or those meeting the broader phenotype of alcohol dependence by COGA criteria. These alcohol dependence phenotypes are largely nested; they are not independent phenotypes, and some of the same SNPs affect risk for more than one of them. There is consistency in the patterns of association among the SNPs in LD with each other.
In summary, we have found evidence for association between GABRR1 and GABRR2 and alcohol dependence in a family-based study, with most of the evidence coming from individuals with early onset of the disorder. This report further supports the role of GABA receptor systems in the involvement of neurotransmission in response to alcohol. It may also provide future directions for functional studies of GABA ρ receptors.
Acknowledgements
The Collaborative Study on the Genetics of Alcoholism (COGA), Co-Principal Investigators B. Porjesz, V. Hesselbrock, H. Edenberg, L. Bierut, includes nine different centers where data collection, analysis, and storage take place. The nine sites and Principal Investigators and Co-Investigators are: University of Connecticut (V. Hesselbrock); Indiana University (H.J. Edenberg, J. Nurnberger Jr., T. Foroud); University of Iowa (S. Kuperman); SUNY Downstate (B. Porjesz); Washington University in St. Louis (L. Bierut, A. Goate, J. Rice); University of California at San Diego (M. Schuckit); Howard University (R. Taylor); Rutgers University (J. Tischfield); Southwest Foundation (L. Almasy). Q. Max Guo is the NIAAA Staff Collaborator. This national collaborative study was supported by the NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA). In memory of Henri Begleiter and Theodore Reich, Principal and Co-Principal Investigators of COGA since its inception; we are indebted to their leadership in the establishment and nurturing of COGA, and acknowledge with great admiration their seminal scientific contributions to the field. SNP genotyping was carried out using the facilities of the Center for Medical Genomics at Indiana University School of Medicine, which is supported in part by the Indiana Genomics Initiative of Indiana University (INGEN®); INGEN is supported in part by The Lilly Endowment, Inc. We thank Gayathri Rajan, Rachel Schultz for expert technical assistance.




