No evidence of an association between two genes, EDN1 and ACE, and childhood-onset mood disorders†
How to Cite this Article: Dempster EL, Kiss E, Kapornai K, Daróczi G, Mayer L, Baji I, Tamas Z, Gadoros J, Kennedy JL, Vetró A, Kovacs M, Barr CL. 2010. No Evidence of an Association Between Two Genes, EDN1 and ACE, and Childhood-Onset Mood Disorders. Am J Med Genet Part B 153B:341–346.
Abstract
Recent evidence supports a pathological link between heart disease and depressive symptoms, suggesting that depression is both etiologic and prognostic to heart disease. Thus, biological molecules which are at the interface between heart and mind are plausible candidate genes for depressive disorders. To investigate this line of enquiry we have investigated two genes, Endothelin 1 (EDN1) and Angiotensin-converting enzyme (ACE) in a family-based sample with childhood-onset mood disorders (COMDs). EDN1 is highly expressed in endothelium where it acts as a potent vasoconstrictor, and is also expressed in the brain where it exhibits neurotransmitter characteristics. ACE acts as a potent vasopressor, and interacts with the hypothalamic–pituitary–adrenocortical (HPA) system, which is often dysregulated in mood disorders. Furthermore, ACE has recently been found to be associated with major depression. Polymorphisms were selected to best capture the genetic variation at the two loci, and to replicate previous associations. The markers were genotyped across EDN1 and ACE in a sample comprised of 382 Hungarian nuclear families ascertained through affected probands diagnosed with a mood disorders before the age of 15. We found no evidence of association between either of these genes and COMD. Consequently, we were unable to support our hypothesis that these two genes, which are involved in both vascular and brain functions are contributing to the susceptibility to mood disorders of children/adolescents. © 2009 Wiley-Liss, Inc.
After depression, coronary heart disease (CHD) is the leading source of disease burden in the western world [Lopez and Murray, 1998]. There is a long established relationship between depression and heart disease, with depressive symptoms appearing to be both etiologic and prognostic to heart disease [McCaffery et al., 2006]. Two independent meta-analyses on available etiological studies [Rugulies, 2002; Wulsin and Singal, 2003] both gave combined odds ratio of depression predicting the onset of coronary artery disease as 1.64. Similarly a meta-analysis on studies examining the effect of depression in patients post-myocardial infarction found that the presence of depression increased the risk of mortality by 2–2.5-fold [van Melle et al., 2004]. Thus, the presence of depressive symptoms increases risk to CHD and worsens prognosis.
A twin study of male twins examined the relationship between depression symptoms and heart disease and found a sizeable genetic correlation of 0.42 between the two disorders [Scherrer et al., 2003], indicating that they share some common genetic predisposing factors. Thus, genes that are expressed in, and whose products have functions in both the vascular and nervous system are compelling candidate genes for mood disorders, as well as cardiac disease.
The vascular endothelium maintains vascular tone by releasing vasoactive substances into the blood vessels. Dysfunction in this cell layer is known as “endothelial dysfunction” and causes alterations in the ability of the endothelial cells to initiate dilation of the blood vessel appropriately to stimuli. This condition is often the first sign of impending heart disease and predates many of the other visual symptoms such as hypertension [Lerman and Zeiher, 2005]. A number of studies have suggested that endothelial function is also impaired in mood disorders [Rajagopalan et al., 2001; Broadley et al., 2002; Sherwood et al., 2005; Rybakowski et al., 2006] and that endothelial dysfunction could be a useful biological trait marker for mood disorders [Rybakowski et al., 2006]. These observations have led to the theory that endothelial dysfunction may be mediating at least some of the comorbidity between these two common disorders.
Endothelin 1 (EDN1) is a peptide with potent vasoconstrictor properties and is upregulated in cardiovascular disease, and is thought to be integral to the development of endothelial dysfunction [Bohm and Pernow, 2007]. The gene encoding EDN1 resides at 6p24-23 and consists of 5 exons covering a total of 6.8 kb [Inoue et al., 1989]. The gene contains a common non-synonymous single nucleotide polymorphisms (SNP; rs5370) which has been associated with blood pressure, but the relationship appears to be complex, such that the association seems to be restricted to overweight individuals [Tiret et al., 1999; Asai et al., 2001; Jin et al., 2003]. Nevertheless, more recently Pare et al. 2007 examined 103 candidate genes for coronary heart disease (CAD) in both a large family-based study, and a case–control sample, drawn from a population that is known to exhibit a founder effect. The non-synonymous SNP, rs5370, in EDN1 emerged as the most significant after correction for multiple comparisons. This association held true in a replication sample from the same geographical area. This study implicates EDN1 as a strong candidate gene for CAD, particularly in females.
EDN1, besides being found in the vascular system, is also expressed in the brain where it exhibits neurotransmitter characteristics. In this role EDN1 has been implicated in both synaptic plasticity [Drew et al., 1998], and nocieption [Hasue et al., 2005], two biological processes that have been implicated in the aetiology of mood disorders [Bair et al., 2003; Normann et al., 2007]. Thus, given the relationship between endothelial dysfunction and mood disorders, in addition to the role of this peptide in the brain, we set out to determine if genetic variations in EDN1 are associated with mood disorders.
In this study we also investigated a second gene, angiotension converting enzyme (ACE), which has been implicated in both coronary heart disease and mood disorders. ACE converts inactive angiotensin I, into a potent vasoconstrictor and aggravator of endothelial dysfunction, angiotensin II, and degrades the vasodilator bradykinin, both of which are important molecules in the maintenance of blood pressure.
The ACE gene is located at 17q23 and consists of two isoforms, 1 and 2, the first is expressed in somatic tissues while the latter is restricted to the testis. The gene is comprised of 25 exons spanning 21 kb. There is a common polymorphic insertion (in/del) of a 287-bp ALU repeat in the gene encoding ACE in intron 16, which is associated with serum levels of ACE [Rigat et al., 1990]. Since its discovery, the ACE in/del polymorphism has been weakly linked to various cardiovascular-related phenotypes [Bleumink et al., 2004; Bondy, 2007]. The ACE gene, however, is in a region of highly conserved linkage disequilibrium (LD) and current opinion favours the presence of other functional polymorphisms in ACE mediating the functional effects associated with the in/del polymorphism [Bleumink et al., 2004].
As angiotensin II contributes to the stress-related activation of the hypothalamic–pituitary–adrenal (HPA)-axis [Armando et al., 2007] ACE has been implicated as a candidate molecule in mood dysregulation. Furthermore, ACE is involved in the metabolism of neurokinins, a family of neurotransmitters including substance P [Yokosawa et al., 1983]. These lines of enquiry have led to the hypothesis that genetic variation in ACE could be associated with depressive disorders. An early study lent support to this hypothesis by reporting an association between the ACE in/del polymorphism and mood disorders [Arinami et al., 1996] leading to a number of replication studies, none of which were able to replicate the original association [Furlong et al., 2000; Meira-Lima et al., 2000; Pauls et al., 2000; Baghai et al., 2001]. Despite the unclear association between this polymorphism and depression, an association has been reported between the in/del and phenotypes related to depressive symptoms, such as cortisol levels in depressed patients [Baghai et al., 2002], and antidepressant efficacy [Baghai et al., 2001, 2004].
Recently, the first comprehensive genetic study on this gene was performed for major depression, which identified a significant association centred on the 5′ end of the ACE isoform 1, and not the in/del polymorphism [Baghai et al., 2006]. Furthermore, the most positive single nucleotide polymorphism (SNP), rs4291, retained its significance in a replication sample, and was found to be associated with ACE levels in response to antidepressant exposure and with cortisol levels.
We endeavoured to replicate the findings of Baghai et al. 2006 by testing for association between ACE 5′ variants and mood disorders. Hence, we investigated polymorphisms in ACE along with polymorphisms in the novel candidate, EDN1, in a large sample of families from Hungary identified through a proband with a diagnosis of a mood disorder before the age of 15.
Diagnosis was ascertained using “The Psychiatric Interview Schedule for Children and Adolescents Diagnostic Version” (ISCA-D) [Sherrill and Kovacs, 2000]. The proband and the parental informant were interviewed individually on two separate occasions approximately 1 month apart by two different child psychiatrists. A consensus diagnosis was then agreed on by two independent child psychiatrists trained in best-estimate diagnosis, more details on the recruitment and assessment of the families in this study are described in detail elsewhere [Burcescu et al., 2006; Liu et al., 2006; Kapornai et al., 2007; Kiss et al., 2007].
This study was approved by the University of Pittsburgh, the Centre for Addiction and Mental Health, Toronto, and all recruitment centres in Hungary. Written informed parental consent and assent from children was obtained for all participants.
Our sample consisted of 382 families with 464 affected children (382 probands and 82 affected siblings, mean age of first onset 10.6 years). The families were recruited from 23 mental health facilities across Hungary. The probands and affected siblings met the DSM-IV criteria for either depressive or bipolar disorder with onset prior to 15 years of age. We also included 6 affected siblings whose onset occurred after 15, but before 18 years of age.
DNA was extracted from whole blood [Miller et al., 1988] and genotyping was performed using the TaqMan 5′ nuclease assay (Applied Biosystems, Foster City, CA) using ABI “assay-on-demand” assays. The TaqMan assays were read on the ABI 7900-HT Sequence Detection System using the Allelic Discrimination End-point Analysis Software version 2.0 (Applied Biosystems, Foster City, CA).
Our data set was free of any detectable Mendelian errors and none of the markers genotyped deviated from the Hardy-Weinberg equilibrium, these quality control checks were performed by the programs PEDSTATS, and MERLIN [Abecasis et al., 2002]. Single marker TDT and haplotype analysis was performed using the TDTphase program from the UNPHASED suite of statistical programs [Dudbridge, 2003]. The haplotype analysis was performed using TRANSMIT [Clayton, 1999] on haplotypes with a frequency greater than 5%. Linkage disequilibrium (LD) between the markers was calculated using Haploview v 3.2 [Barrett et al., 2005].
To minimise the number of markers required “tag SNPs” were chosen, along with SNPs identified from previous reports as being associated to disease or function. EDN1: Tag SNPs were chosen by the HAPLOVIEW program using the HAPMAP build from October 2005, three tag SNPS were highlighted, rs14760, rs5369 and rs207194 for analysis. A further two SNPs, rs10478694 and rs5370, were selected based on previous association and functionality reports [Popowski et al., 2003; Pare et al., 2007]. Moreover, by adding the five chosen EDN1 SNPs in to the SNP tagging selection program Tagger and by using the latest January 2007 HAPMAP build, we can confirm that these 5 SNPs give adequate coverage of the gene with high LD across the region (D′ > 0.85)
ACE: Tag SNPs were picked by the Tagger program using the HAPMAP build from March 2008. The selection criteria of r2 = 0.8 and minor allele frequency (MAF) = 0.1, resulted in the selection of 5 tag SNPs; rs4295, rs4309, rs4311, rs4305 and rs4351. All these SNPs apart for one, rs4305, were chosen for analysis. Rs4305 was not genotyped for two reasons, first it was negative in the study by Baghai et al. 2006, and second there was not an available assay. Instead, we replaced this SNP with the most positive SNP from the Baghai et al. 2006 study, rs4291.
EDN1: Five SNP markers were genotyped across the EDN1 gene, covering a 5.5 kb region. Four of the SNPs were in tight LD, i.e. D′ values did not fall below 0.96 for any given marker combination (Fig. 1).
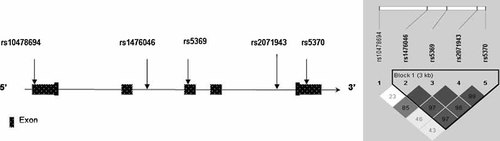
Diagram (not to scale) illustrating the location of the five SNPs genotyped across the EDN1 gene.
The single marker TDT analysis for these five markers is shown in Table I. There was no evidence of association between any of the SNPs in EDN1 and COMD.
| EDN1 SNPs | SNP type | dbSNP build 126 | Location in EDN1 | MAF | T | NT | χ2 | P value |
|---|---|---|---|---|---|---|---|---|
| rs10478694 | (In/del “A”) | Chr6:12398718 | 5′UTR | (A ins) 0.31 | (A del) 137 | 145 | 0.23 | 0.63 |
| rs1476046 | (G>A) | Chr6:12401207 | Intron 2 | (A) 0.23 | (G) 126 | 120 | 0.15 | 0.70 |
| rs5369 | (G>A) | Chr6:12402244 | Exon 3 (synonymous) | (A) 0.13 | (G) 78 | 94 | 1.49 | 0.22 |
| rs2071943 | (G>A) | Chr6:12403800 | Intron 4 | (A) 0.21 | (G) 126 | 117 | 0.33 | 0.56 |
| rs5370 | Lys198Asn (G>T) | Chr6:12404241 | Exon 5 (non-synonymous) | (T) 0.21 | (G) 124 | 117 | 0.20 | 0.65 |
Haplotype analysis, using the four markers in high LD with one another (rs14760–rs5369–rs207194–rs5370) revealed three haplotypes over 5%. The most common haplotype was at a frequency of 63%, with less common haplotypes present at 20%, and 13%. The haplotype association test was negative, with no evidence of bias transmission to affected children for any of the EDN1 haplotypes (not shown).
ACE: Five SNP markers were genotyped across the ACE gene, covering a 15.5 kb region. These SNPs are located in a region of high LD, as evidenced by pair-wise D′ LD values not falling below 0.91 (see Fig. 2).
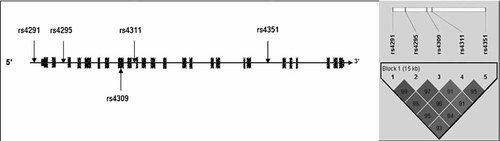
Diagram (not to scale) illustrating the location of the five SNPs genotyped across the ACE isoform 1.
The single marker TDT analysis for these five markers is shown in Table II. Again, there was no evidence of association between any of the SNPs in ACE and COMD.
| ACE SNP | SNP type | dbSNP build 125 | Location in ACE isoform 1 | Replication of Baghai et al. or Tag SNP | MAF | T | NT | χ2 | P value |
|---|---|---|---|---|---|---|---|---|---|
| rs4291 | (A>T) | Chr17:58907926 | Promoter | Replication (T allele has previously found to be associated) | (T) 0.41 | (A) 179 | 163 | 0.74 | 0.38 |
| rs4295 | (G>C) | Chr17:58910030 | Intron 2 | Tag and Replication (G allele has previously found to be associated) | (C) 0.41 | (G) 165 | 178 | 0.05 | 0.48 |
| rs4309 | (C>T) | Chr17:58913655 | Exon 8 (Synonymous) | Tag SNP | (T) 0.42 | (C) 174 | 194 | 1.08 | 0.29 |
| rs4311 | (C>T) | Chr17:58914495 | Intron 9 | Tag and Replication | (T) 0.49 | (C) 195 | 167 | 2.17 | 0.14 |
| rs4351 | (G>A) | Chr17:58923464 | Intron 19 | Tag SNP | (G) 0.46 | (G) 185 | 160 | 1.81 | 0.18 |
Haplotype analysis revealed two common haplotypes which both had a frequency of 39% and one less frequent haplotype at 7%. No association was found between the ACE haplotypes and COMD (not shown).
In this study we have examined two genes, EDN1 and ACE, which were chosen for investigation based on the evidence that mood disorders and vascular disease share a genetic aetiology and both these genes have functions related to both disorders. However, we failed to find any evidence of association between either genes and COMD in this sample.
This is the first genetic study, to our knowledge, of the gene EDN1 and mood disorders. It was chosen based on recent literature that observed an increase in endothelial dysfunction in individuals diagnosed with depression. However the outcome of this study using these markers did not support this gene as a major contributor to genetic vulnerability to COMD in this sample. Although, it is possible that this gene predisposes to endothelial dysfunction, which in turn, increases risk for depression through an alterative mechanism. Future work should examine this gene in relation to endothelial dysfunction in depressed populations.
ACE, on the other hand, has been previously associated with mood disorders and has been postulated to play a role in the HPA-axis, a system whose dysregulation is thought to be integral to the depression phenotype. The lack of association was not predicted given that we have typed the same markers that were found to be significant in a recent case-control study by Baghai et al. 2006, including rs4295 which gave an odds ratio of 1.91. Common explanations for lack of consistency between association studies include inadequate power, yet our sample which was comprised of nearly 400 families, had 98% power to detect an odds ratio of 1.9, as calculated by the program Quanto (http://hydra.usc.edu/gxe). An alterative explanation to our inability to replicate the previous report of association between ACE gene variants and depressive symptoms may lie in differences between the ascertainment criteria of the two samples. In the Baghai et al. 2006 study co-morbidity of heart disease was not an exclusion criterion for their major depression cases, and moreover they actually found an association between their most positive ACE marker rs4291 and hypertension. This indicates that rs4291 is a risk factor for both disorders, implying that genetic variation in ACE is a risk factor for the combined phenotype of depressive and cardiac symptoms. Therefore, perhaps our lack of replication with ACE is indicative of less individuals with a “combined genetic risk” to mood disorders and heart disease in our sample, and obviously given our samples average age at assessment (11.7 years) there is no way of testing this as we have no way of predicting which children will develop heart disease in the future. Moreover, the relationship of depression and cardiac illness maybe confined to those with a later age of onset of depression symptoms. Age of onset has been suggested to be source of genetic variation in mood disorders, thus it is pertinent that there could be different genetic, and environmental factors, which influence the onset and nature of co-morbidities such as cardiac disease with depression depending when in life the onset of mood disorders first occurs.
To conclude we have examined two genes, EDN1 and ACE, under the premise that genes involved in mechanisms applicable to both heart disease and depressive disorders would be strong candidates for COMD, the finding of such an association would provide sound evidence of the existence of a genetic link between heart disease and depression. We were, however, unable to find any evidence of association between the two genes and COMD in this study, but we cannot rule out involvement of either gene between later onset depression and cardiac illness.




