A narrow and highly significant linkage signal for severe bipolar disorder in the chromosome 5q33 region in Latin American pedigrees†
How to cite this article: Jasinska AJ, Service S, Jawaheer D, DeYoung J, Levinson M, Zhang Z, Kremeyer B, Muller H, Aldana I, Garcia J, Restrepo G, Lopez C, Palacio C, Duque C, Parra M, Vega J, Ortiz D, Bedoya G, Mathews C, Davanzo P, Fournier E, Bejarano J, Ramirez M, Araya Ortiz C, Araya X, Molina J, Sabatti C, Reus V, Ospina J, Macaya G, Ruiz-Linares A, Freimer NB. 2009. A Narrow and Highly Significant Linkage Signal for Severe Bipolar Disorder in the Chromosome 5q33 Region in Latin American Pedigrees. Am J Med Genet Part B 150B:998–1006.
Abstract
We previously reported linkage of bipolar disorder to 5q33-q34 in families from two closely related population isolates, the Central Valley of Costa Rica (CVCR) and Antioquia, Colombia (CO). Here we present follow up results from fine-scale mapping in large CVCR and CO families segregating severe bipolar disorder, BP-I, and in 343 population trios/duos from CVCR and CO. Employing densely spaced SNPs to fine map the prior linkage peak region increases linkage evidence and clarifies the position of the putative BP-I locus. We performed two-point linkage analysis with 1134 SNPs in an approximately 9 Mb region between markers D5S410 and D5S422. Combining pedigrees from CVCR and CO yields a LOD score of 4.9 at SNP rs10035961. Two other SNPs (rs7721142 and rs1422795) within the same 94 kb region also displayed LOD scores greater than 4. This linkage peak coincides with our prior microsatellite results and suggests a narrowed BP-I susceptibility regions in these families. To investigate if the locus implicated in the familial form of BP-I also contributes to disease risk in the population, we followed up the family results with association analysis in duo and trio samples, obtaining signals within 2 Mb of the peak linkage signal in the pedigrees; rs12523547 and rs267015 (P = 0.00004 and 0.00016, respectively) in the CO sample and rs244960 in the CVCR sample and the combined sample, with P = 0.00032 and 0.00016, respectively. It remains unclear whether these association results reflect the same locus contributing to BP susceptibility within the extended pedigrees. © 2009 Wiley-Liss, Inc.
INTRODUCTION
Despite overwhelming evidence from family, twin and adoption studies that bipolar disorder (BP, MIM %125480) has a strong heritable component, efforts to identify sequence variants predisposing to this illness have so far proven unsuccessful. Linkage and association studies have implicated multiple putative chromosomal locations for BP susceptibility loci [Craddock and Forty, 2006], but none of these findings have been convincingly replicated. Loss of power due to both phenotypic heterogeneity and genetic complexity probably account for the equivocal results of BP mapping studies to date. One approach to increase the power for BP mapping studies is to investigate relatively homogeneous samples, either by including only individuals with a narrow phenotype definition or by focusing on subjects from genetically restricted population isolates. In such populations, increased inbreeding and rapid population growth in relative isolation from other populations may lead to reduced genetic complexity [Peltonen et al., 2000] and variants associated with risk for common diseases may be shared by a larger fraction of affected individuals than in outbred populations. Additionally, in comparison with outbred populations, recent founder populations typically display more extensive linkage disequilibrium (LD), which may provide the power to detect associations with fewer markers [Service et al., 2006a].
We previously performed genome-wide linkage studies of severe bipolar disorder (BP-I) in a series of extended pedigrees from the Central Valley of Costa Rica (CVCR) and the province of Antioquia in Northwest Colombia (CO). Both populations were founded mainly by admixture between small numbers of Amerindian women and Spanish men in the 16th–17th centuries, and since that time they have grown rapidly (over 1,000-fold increase in size) in relative isolation [Carvajal-Carmona et al., 2003]. The close genetic relatedness of these two population isolates has been shown by a variety of measures obtained using mitochondrial, Y-chromosome, and autosomal markers (microsatellites and SNPs) [Carvajal-Carmona et al., 2003; Service et al., 2006a]. The genome scan that we conducted in three large CVCR pedigrees and 14 CO pedigrees provided convergent linkage evidence, at a genome-wide significance threshold, for a BP-I susceptibility locus in the 5q31-34 chromosomal region [Herzberg et al., 2006]. We then narrowed the region down further to ∼10 cM around the marker D5S2049 using microsatellites spaced at 1–2 cM [Herzberg et al., 2006]. The implicated region (5q33-34) has shown evidence of linkage in multiple previous genetic studies of schizophrenia and mood disorders, suggesting strongly that it may contain a locus predisposing to severe psychiatric illness [Park et al., 2004; Etain et al., 2006; Venken and Del-Favero, 2007].
In the current study, we followed up the significant linkage signal at 5q33-34 previously showing the greatest genome-wide evidence of linkage for BP-I in the CVCR and CO families. We employed a dense SNP map, to attempt to narrow the linkage signal and possibly increase its intensity. Next, we performed a test for association in the presence of linkage in the families in order to identify more precisely the localization of the variant(s) responsible for the linkage signal.
The strong linkage signal observed in these pedigrees is consistent with the action of highly penetrant mutations that greatly increase the risk of disease in carriers. Such variants, while typically infrequent, could also contribute to population disease risk, as could more common variants at the same locus. To additionally refine the linkage signal obtained from the pedigree study and to determine the role, in the population, of loci responsible for familial BP-I aggregation, we used the same dense set of SNPs and tested for association in population samples. We further investigated possible overlap between association signals from family and population samples. Here we present highly significant linkage results delineating a narrow chromosomal location within the 5q33 region, which contains promising functional candidate genes for BP-I. Follow up of the linkage signal with association analysis in the population trio sample produced association signals at three marker loci, but they did not overlap clearly with the linkage signal in the pedigrees. Non-overlapping linkage and association signals in the 5q33 region may suggest either a different genetic basis of BP-I in pedigree and population samples, or allelic heterogeneity between the different study samples.
MATERIALS AND METHODS
Subjects
All subjects possibly affected with BP-I were interviewed using the Diagnostic Interview for Genetic Studies (DIGS, version 3) [Nurnberger et al., 1994]. The medical records and DIGS were reviewed by two psychiatrists who independently diagnosed each subject. This best estimate procedure has been described in detail [Escamilla et al., 1996; Freimer et al., 1996]. Only subjects with a best estimate diagnosis of definite BP-I were considered affected for purposes of linkage or association analysis.
Pedigree samples used in this study come from three large families from the CVCR and 14 intermediate size pedigrees from CO. Extensive genealogic information documents the descent of most individuals in these pedigrees form the CVCR and CO founding populations. The average pedigree size was 16 persons (ranging from 6 to 41), with an average of five BP-I affected individuals per pedigree (ranging from 2 to 13). The largest CVCR pedigree, CR201, includes 25 individuals diagnosed with BP-I [Service et al., 2006b]. Two other CVCR pedigrees, CR001 and CR004, are related by several inbreeding loops and altogether include 30 individuals affected with BP-I [Freimer et al., 1996]. The CO pedigrees contain 3–13 BP-I cases per family and altogether include 85 BP-I patients [Herzberg et al., 2006].
The sample for association analyses comprised independently ascertained BP-I affected subjects and their parent(s). These individuals were recruited, without consideration of family history of psychiatric disorders, from patients admitted with a diagnosis of BP-I to psychiatric hospitals in CVCR and CO. Inclusion in the study was restricted to individuals with a best estimate diagnosis of BP-I, at least one psychiatric hospitalization and age of onset ≤50 years old, at least one parent available for genotyping, and at least six of eight great-grandparents of CVCR or CO ancestry. This genealogic screen ensures that most of the ancestry of the included individuals derives from the founding populations of the two isolates. A total of 249 patients from CVCR and 110 patients from CO met these criteria. In the CVCR sample set, 101 patients had both parents available for genotyping (trios), and 148 had only one parent available for genotyping (duos). The CO sample set consisted of 54 trios and 56 duos available for genotyping.
DNA Samples
Genomic DNA samples were obtained from whole blood with alcohol and salt precipitation using an automated Autopure LS system (Qiagen Sciences Inc., Germantown, MD) and Puregene chemistry (Gentra) or a standard phenol/chlorophorm extraction procedure. The quantity of double-stranded template in total genomic DNA or, if the amount of genomic DNA was very limited, a product of whole-genome amplification (REPLI-g Kit; Qiagen) was quantified by a Quant-iT PicoGreen assay (Invitrogen, Carlsbad, CA) and all samples were normalized to a concentration of approximately 50 ng/µl.
SNP Marker Selection
SNPs for linkage and association fine mapping were selected in the ∼9.3 Mb region centered around D5S2049, the marker showing the strongest evidence of linkage to BP-I in our previous study [Herzberg et al., 2006]. This ∼9.3 Mb region is bounded by markers D5S410 and D5S422, and delineated linkage results within one LOD score unit of the score for D5S2049 (Fig. 1A). To select informative SNPs, which capture genetic variation in the region most efficiently, and to avoid analyzing redundant information from SNPs in strong LD, we selected haplotype tagging SNPs (tSNPs). Using the pair-wise tagging algorithm incorporated in Haploview software version 3.2 [Barrett et al., 2005], we selected tSNPs based on the genotype data from the CEU individuals of Northern and Western European ancestry available from the HapMap Project (http://www.hapmap.org/); at the time of SNP selection for this project, this represented the reference data set most relevant to the CVCR and CO populations. We aimed to assay the 5q33 region using a single SNP array, and to provide enhanced coverage in low LD regions and in segments containing previously proposed candidate genes for major psychiatric disorders. We further assumed that the extensive LD observed in both populations in prior studies [Service et al., 2006a] would enable us to capture most of the LD in the region using somewhat fewer SNPs than required in outbred populations.
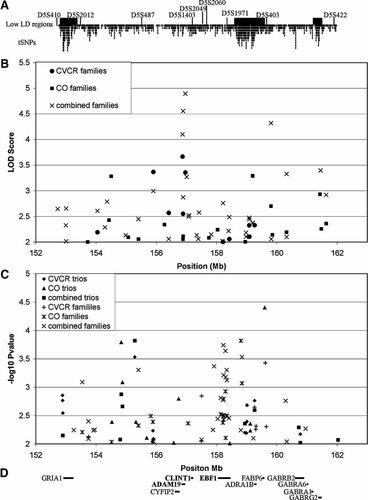
Results of two-point linkage and association with BP-I in families and trios from Costa Rica and Colombia using densely spaced SNP markers. Distribution of microsatellite markers analyzed previously [Herzberg et al., 2006] and tested SNPs (black vertical bars) are shown with respect to localization of low LD regions (gray horizontal bars), which were covered with increased density of SNPs in 5q33 (A). Two-point linkage analysis in BP-I families from CVCR (circle), CO (square), and combined results (×) is shown in panel (B). Tests of association given linkage were done using frequencies estimated from the corresponding trio sample. The LOD score (Y-axis) is plotted against of the physical position in megabases (Mb) of the corresponding SNPs. For clarity, only LOD scores greater than 2 are shown. Panel (C) shows association in presence of linkage in CVCR families (cross), CO families (star), and combined results (×); and association in population trios from CVCR (diamond), CO (triangle), and in combined trio sample (square) as −log10 P-value (Y-axis) plotted against physical position in Mb of the corresponding SNP. For clarity, only results with −log10 P-value greater than 2 are plotted. Positions of the RefSeq genes in the investigated region are shown in panel (D). The most promising positional and functional candidate genes in the region are shown in boldface.
For the majority of the investigated region (∼8 Mb), the Phase I release #16 HapMap data (CEU) was used to select tSNPs. To provide SNP density adequate to the extent of LD in the targeted genomic region, we constructed an LD map [Maniatis et al., 2002] based on these data. This map indicated three low LD regions, defined as a gap of at least 2.5 LD units between consecutive SNPs as plotted relative to NCBI Build 35 in Figure 1A. Since markers located in very low LD regions may have limited power to detect association [Service et al., 2006a], tSNPs were selected with increased density in these regions (∼1.3 Mb) using an approximately threefold denser map from Phase II release #20 (CEU) and their genomic positions converted to NCBI Build 35 (hg17) are shown in Figure 1A. The tSNPs with poor Illumina quality scores, which predicted low confidence genotype calls, were replaced with other tSNPs, which could be pooled in the Illumina assay. Additionally, 15 common SNPs were added within or in the proximity of the genes for which function can be potentially related to BP. Altogether 1,134 SNPs were selected for this study. Over the entire region, the SNPs chosen all displayed minor allele frequencies in CEU of >0.05 and showed a minimum r2 of 0.73 with tagged SNPs.
SNP Genotyping
SNP marker loci were genotyped using the Golden Gate Assay employing allele-specific extension and ligation methodology followed by universal-primer PCR amplification reactions (Illumina, San Diego, CA, http://www.illumina.com). Genotyping reactions were performed using 250 ng of normalized DNA sample as a template. The samples were processed on the Illumina BeadLab 1000 platform according to the manufacturer's protocols [Fan et al., 2003]. Genotypes were clustered automatically using GeneCall program version 6.0.7 (Illumina) and then confirmed by visual inspection and if necessary corrected manually. Gene calls were extracted using the gene calling programs and GTS Reports (Illumina). The quality GenCall Scores (GCS) were used to identify and discard samples, markers and individual genotypes below 0.25 thresholds.
QC of Genotyping Data
Genotype data quality was evaluated using the trio samples. HWE was assessed using an exact test. Markers with P < 0.001 from this test were flagged for further inspection. Mendelian segregation in the trios was assessed. Markers with a different rate of missing data in CVCR and CO were also flagged for further inspection, as were markers with few homozygotes or few heterozygotes. For all flagged markers, clustering plots were manually inspected. If the scoring appeared to be questionable, markers were rescored and retested. If problems remained, they were discarded from further analyses. A total of 16 affected individuals (4 CVCR and 12 CO) were removed from the association analyses due to excessive Mendelian errors (see Results Section). If the scoring was not questionable, markers out of HWE (five markers) or with excessive Mendelian errors (four markers with errors in >5% of trios) were discarded. Both sample and SNP genotype completeness and quality checks are described in detail in Supplementary Materials. Evaluation of possible copy number variations was conducted using PennCNV [Wang et al., 2007], which is based on a Hidden Markov Model (HMM) that utilizes the log R ratio, a measure developed by Illumina as a normalized signal intensity, and B allele frequency. Prior to running PennCNV data were pre-processed to eliminate systematic fluctuations in the log R ratio. Identified CNV variants were visually inspected.
Statistical Analysis
Two-point parametric linkage analysis in the pedigree samples was performed using the linkage option in Mendel [Lange et al., 2001]. SNP allele frequencies were estimated using parents of the BP-I trio association samples. There were 343 parents genotyped from CVCR and 148 parents genotyped from CO. Parametric analysis with Mendel was performed under the model previously employed for CVCR families [McInnes et al., 1996]. This model assumes a causative allele with frequency 0.003. The penetrance in individuals homozygous for the normal allele was set to 0.01, for the heterozygote was 0.81, and for individuals homozygous for the causative allele penetrance was set to 0.90. This nearly dominant model is consistent with the epidemiological data showing a world-wide disease prevalence of ∼1.5%.
Since the pedigrees were too large and too complex for multipoint analysis with all markers tested in the region, 25 markers were chosen for this analysis based on low LD between each other and high MAF (greater than 0.3). Multipoint linkage analysis with selected markers was performed using SimWalk2 based on the genetic position in the recombination maps generated by HapMap.
Association analysis suitable for the pedigrees was conducted using the association given linkage option in Mendel [Cantor et al., 2005] using the same allele frequency estimates as for linkage analysis.
A two-point test of association was performed using Transmit [Clayton, 1999] for both duos and trios. As a test of association in the Transmit analysis, we used the asymptotic chi-squared test. This is a test with 1 df for excess transmission of an allele.
RESULTS
We saturated the 9.3 Mb region around D5S2049 with 1,134 SNP markers. A total of 1,082 SNPs, which passed quality control checks, were used in further statistical analyses in 17 pedigrees and 343 trios with BP-I probands from the CVCR and CO populations. Details on completeness and quality control of the SNP markers and trio samples are presented in Supplementary Materials. No reliable copy number variation was identified in this sample (data not shown).
Linkage and Association in Presence of Linkage in Families
Two-point parametric linkage results for the 1,082 markers that passed quality checks are presented in Figure 1B. We observed a highly significant LOD score of 4.9 at the rs10035961 locus (156.9 Mb), in the combined CVCR and CO pedigrees. Additionally, two other SNPs (rs7721142 and rs1422795) in this region also displayed a combined LOD score greater than 4. The location of this linkage peak was not sensitive to alternative specifications of the genetic model (data not shown).
Although the pedigree samples from both populations contribute to this signal in the central part of the investigated region, the major contributors to this result are large CVCR families, which, on their own, produce a significant linkage signal in this region. The second most significant location in the analysis of all families is highlighted by rs4921283 (159.8 Mb) and this signal comes mostly from the CO pedigrees.
A LOD score greater than 3 was obtained for three SNPs, rs253602, rs1422795, and rs10035961, in CVCR families analyzed alone (maximum of 3.7 at rs1422795). These markers are located in about 1.1 Mb of the central part of the investigated region around nucleotide position 157 Mb (Table I). A suggestive LOD score was also observed for rs7721195 in this region. Three other SNPs, rs6863955, rs10036026, and rs9313827, displayed a LOD score greater than 3 in CO families analyzed alone. These SNPs were in two distinct locations, at 154.5 and 159.2 Mb. Locus rs183294 at 161.4 Mb reached a LOD score of 2.9 in the CO families.
| Marker | Physical position (nt)a | LOD score, CVCR | LOD score, CO | Combined LOD score |
|---|---|---|---|---|
| rs6863955 | 154504103 | 0.40 | 3.28 | 1.78 |
| rs253602 | 155898665 | 3.37 | 0.57 | 2.99 |
| rs1422795 | 156868942 | 3.67 | 1.15 | 4.10 |
| rs7721142 | 156884986 | 2.55 | 2.05 | 4.56 |
| rs10035961 | 156962973 | 3.36 | 1.69 | 4.89 |
| rs7713029 | 157008023 | 1.72 | 1.72 | 3.27 |
| rs10036026 | 159195539 | 0.17 | 3.29 | 2.37 |
| rs9313827 | 159196375 | 0.17 | 3.29 | 2.38 |
| rs4921283 | 159803189 | 1.76 | 2.70 | 4.32 |
| rs2176297 | 160321177 | 1.18 | 2.19 | 3.33 |
| rs209353 | 161449637 | 1.55 | 1.85 | 3.40 |
- a NCBI Build 35.
Using the CO and CVCR families, we tested for linkage in the presence of heterogeneity for all SNPs in Table I. In all tests there was no evidence for heterogeneity between CO and CVCR families, and the linked proportion was 1.0.
Multipoint analysis with Simwalk2 was conducted to further refine the obtained linkage signal, but the results of multipoint mapping implicated the entire region and did not further clarify the linkage peak from the two-point analysis (data not shown).
We also conducted an analysis testing association given linkage in the pedigrees (Fig. 1C). While we observed no associations that met our significance threshold (P = 0.00003, considering a correction for all 1,082 tested SNPs), a group of markers located around 158.3 Mb showed association signals in the combined CVCR and CO families (eight SNPs) and in CO families only (two SNPs) (Table II). The highest combined signal for association in CVCR and CO families was obtained for three of these markers (rs31199, rs6556373, and rs10515794 with respective P-values of 0.00020, 0.00025, and 0.00032) in a ∼0.6 Mb region between 158.2 and 158.8 Mb. This region contains also a marker rs2421182, which shows the highest level of significance in CO families (P = 0.00016).
| Marker | Physical position (nt)a | CVCR triosb | CO triosb | Combined triosb | CVCR families c | CO families c | Combined familiesc |
|---|---|---|---|---|---|---|---|
| rs10515705 | 153540472 | 0.79433 | 0.79433 | 0.79433 | 0.50119 | 0.00631 | 0.00079 |
| rs267015 | 154836363 | 0.25119 | 0.00016 | 0.00126 | 0.39811 | 0.79433 | 0.50119 |
| rs977776 | 154875106 | 0.19953 | 0.00079 | 0.00200 | 0.39811 | 0.79433 | 0.50119 |
| rs244960 | 155283839 | 0.00032 | 0.19953 | 0.00016 | 0.25119 | 0.05012 | 0.12589 |
| rs6556352 | 155404292 | 0.19953 | 1 | 0.31623 | 0.05012 | 0.00501 | 0.00050 |
| rs2963426 | 157914908 | 1 | 0.12589 | 0.31623 | 0.19953 | 0.01585 | 0.00040 |
| rs31199 | 158222673 | 0.63096 | 0.25119 | 0.79433 | 0.05012 | 0.00316 | 0.00020 |
| rs9313794 | 158232438 | 0.79433 | 0.31623 | 0.63096 | 0.07943 | 0.00398 | 0.00063 |
| rs6556373 | 158292054 | 0.79433 | 0.63096 | 0.63096 | 0.25119 | 0.00126 | 0.00025 |
| rs4921120 | 158301973 | 1 | 1 | 1 | 1 | 0.00794 | 0.00063 |
| rs6556380 | 158333106 | 0.63096 | 0.19953 | 0.79433 | 0.06310 | 0.00316 | 0.00050 |
| rs12153593 | 158342659 | 0.79433 | 0.79433 | 1 | 0.10000 | 0.00316 | 0.00079 |
| rs2421182 | 158805218 | 0.79433 | 0.79433 | 0.79433 | 1 | 0.00016 | 0.00631 |
| rs10515794 | 158840805 | 0.31623 | 1 | 0.39811 | 0.19953 | 0.00079 | 0.00032 |
| rs12523547 | 159595302 | 0.79433 | 0.00004 | 0.02512 | 0.15849 | 0.79433 | 0.12589 |
| rs3749799 | 159615355 | 0.39811 | 0.01000 | 0.39811 | 0.00040 | 0.79433 | 0.03981 |
- a NCBI Build 35.
- b Association in trios calculated with Transmit.
- c Association given linkage in pedigrees calculated with Mendel.
Association Analysis in Population Samples
The same set of SNP markers was tested for association in trio samples from both populations. The association test used, Transmit, is a family-based test and is robust to population stratification even in the case where one parent is missing data. Allele frequencies in CO and CVCR are very similar for the SNPs typed in 5q33 (Fig. 2), indicating minimal stratification in the combined sample. Using Transmit, we observed marginal association signals in two regions. The strongest association (P = 0.00004) was obtained for rs12523547 (159.5 Mb) in the CO population but no other nearby marker showed a comparable signal. An additional region showing association was demarcated by markers rs244960 (155.3 Mb) reaching a significance level of P = 0.00032 in the combined trio samples, with a major contribution from the CVCR samples, and rs267015 (154.8 Mb) reaching a significance level of P = 0.00016 in the CO samples. This region is approximately 2 Mb upstream from the narrow linkage peak obtained in the families.
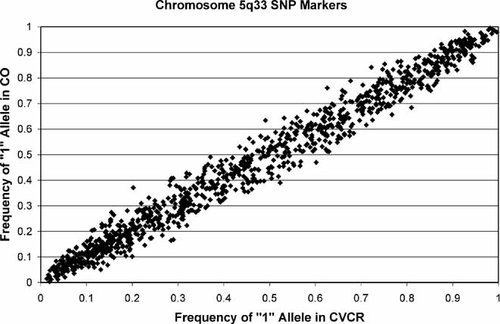
Comparison of allele frequencies in the CR and CVCR population samples. Allele frequencies were estimated from parental data. Presented is the frequency of the “1” allele at each marker.
DISCUSSION
In this article, we present results of our studies aiming to refine a previously reported linkage peak at 5q31-33 for BP-I in pedigrees from population isolates from CO and CVCR. We hypothesized that variants associated with the disease in the population might be located at the same locus as risk alleles responsible for familial aggregations of the disease and that their possible co-localization might be helpful in identifying the disease predisposing locus. Therefore, we followed up the BP-I linkage signal by genotyping the same markers in both large families and population trio samples.
We performed high-resolution fine mapping in these samples taking advantage of over 1,000 SNPs spaced at about 8.5 kb distance across the previously identified linkage region. While SNPs are on average less informative than microsatellite markers, the use of large numbers of SNPs provides the opportunity to identify stronger linkage evidence than is observed using a sparser set of microsatellites [Freimer et al., 2007]. In the current study, employing a dense SNP map, we observed an increase in statistical evidence for linkage in the region. Previously, we obtained a peak multipoint NPL score of 4.4 (Simwalk2) and two-point parametric linkage of 2.5 from microsatellite markers in these families [Herzberg et al., 2006]. Parametric two-point linkage analysis with the same model and a dense set of SNP markers resulted in a total of seven SNPs with LOD scores exceeding 3, among them marker rs10035961 giving the highest LOD score of 4.9. Additionally, using densely spaced SNP markers allowed us to narrow down considerably the likely region of interest. From SNPs in the region with LOD scores greater than 3, at least four SNPs (rs1422795, rs7721142, rs10035961, and rs7713029) covering a 140 kb region, overlap the initial microsatellite linkage peak.
In spite of the increased magnitude and resolution of the linkage signal, the association results do not contribute additional information towards identification of BP-I susceptibility variants in this region. The low association signal most likely reflects the relatively small sample size, even combining the two populations. Additionally, the SNP density used may have provided inadequate coverage of the region, even considering the high LD that characterizes these population isolates.
It is well known that population stratification can result in spurious association findings. Our association samples come from two populations, CO and CVCR, which have experienced similar degrees of admixture at the time of European settlement. Mitochondrial and Y chromosome [Carvajal-Carmona et al., 2003], as well as analysis of autosomal SNP markers [Service et al., 2006a], suggest that these populations are genetically similar and share a common demographic history. The very similar allele frequency distributions in the two populations in the 5q region is in accord with these previous findings. Substantial stratification is also unlikely within each population, given the restricted immigration to both populations subsequent to the initial admixture of Europeans and Amerindians within the first century after the founding of each population. Furthermore, the use of the Transmit program for the association analyses provides an additional check against unexpected substructure that would affect association results. It is also unlikely that stratification accounts for different results between the pedigree and population samples; in both CO and CVCR extensive genealogies document the descent of the pedigrees nearly exclusively from the founding populations.
The narrow linkage peak does not show clear co-localization with the most meaningful association signals in the pedigrees and trios. In the pedigrees, this observation may suggest the contribution of more than one disease-related locus in the region or the presence at a single locus of multiple susceptibility alleles in these families, particularly those that are very large. In the presence of locus and allelic heterogeneity, different families may still be contributing to the linkage signals. Linkage is robust to allelic heterogeneity, whereas association tests are not. Multiple variants in the linkage region would be difficult to detect in this small association sample. Such a scenario could explain the lack of association signal in the immediate vicinity of the linkage peak, and detecting such variants in BP-I individuals in the CO and CVCR populations would likely require direct sequencing.
An alternative explanation for the lack of overlap between the association signals that we observed in the trios and the narrow linkage peak observed in the pedigrees is that predisposition to BP-I in heavily loaded families and population risk of BP-I are conferred by different types of genetic variants, even within these population isolates. Different genes are known to contribute to familial and sporadic cases of other common diseases such as Alzheimer's disease where the amyloid precursor and presenilin genes are responsible mostly for rare forms of familial Alzheimer disease while other loci, such as ApoE, confer increased risk for the majority of sporadic cases, which are characterized by a complex polygenic background [St George-Hyslop, 2000].
The association results may suggest two loci, EBF1 and FABP6, as interesting candidate genes. Association in the presence of linkage highlights the EBF1 gene (early B-cell factor, MIM *164343) whose intron 4 contains five SNPs (rs31199, rs9313794, rs6556373, rs4921120, rs6556380, and rs12153593) showing association of P < 0.001. The role of the EBF1 gene as a transcription factor essential to the differentiation of striatonigral neurons [Lobo et al., 2006] and the presence in the gene region of multiple conservative elements, makes it an interesting candidate as well. The strongest association signal in the population (P = 0.00004) was found at rs12523547 located in intronic region of the FABP6 gene (gastrotropin, MIM *600422).
Even though association results do not provide additional evidence for localization of the BP-I locus, the highly significant linkage results encourage further studies focused on the 5q33 region in the pedigrees contributing to the signal. The narrow linkage region encompasses two strong candidate genes, the function of which could be involved in the disease phenotype. ADAM19 (ADAM metallopeptidase domain 19, MIM *603640) and CLINT1 (clathrin interactor 1, MIM #181510) are the genes nearest the implicated region. ADAM19, which is directly pinpointed by the presence of an exonic SNP significantly linked to BP-I (rs1422795, LOD = 4.1), is an interesting functional candidate due to its role in the proteolytic processing of neuregulin 1 [Yokozeki et al., 2007], which is a key element of the neuregulin-erbB signaling pathway, putatively impaired in psychiatric diseases [Roy et al., 2007]. The second candidate gene, CLINT1, is suggested as a candidate for vulnerability to psychopathology by its association with schizophrenia in UK, Chinese, and Latin American populations [Pimm et al., 2005; Liou et al., 2006; Tang et al., 2006; Gurling et al., 2007; Escamilla et al., 2008]. Epsin 4 which is a protein product of the CLINT1 gene was shown to be involved in endocytotic internalization, regulation of inositol phospholipid levels, membrane structure, and trafficking through direct binding to membrane clathrin and other associated proteins, such as AP-1 and AP-2 [McPherson and Ritter, 2005]. We consider CLINT1 the strongest candidate on the basis of its involvement in processes which are linked to lithium's therapeutic action as a mood stabilizer such as regulation of phosphoinositol turnover and its effects on phosphoinositide 3-kinase and protein kinase C [Sato et al., 1996; McPherson and Ritter, 2005].
Among the several complementary analyses that we applied to fine map the BP-I locus in the 5q33-34 region, the linkage analysis generated the most compelling results. These analyses are less affected by SNP selection and limited sample size than the other analyses performed, and produced a highly significant signal, consistent across several markers and highlighting a very narrow candidate region. The small size of the implicated region makes it feasible to attempt to identify variants contributing to familial BP by applying a candidate gene or region-wide re-sequencing strategy, supplemented by detection of large insertions/deletions and copy number variation. In this approach aimed at detection of deleterious variants segregating in the large pedigrees, it is expected that identification of disease predisposing variants among other sequence variation in the region will be based on a clear functional effect of such variants.
Acknowledgements
This work was partly funded by Universidad de Antioquia (CODI), the Wellcome Trust (grant 086052), a NARSAD young investigator award to A.R.-L., and NIH grants R01 MH 049499 and K02 MH 001375 to N.F. We would like to thank the members of the families for their participation as well as Alfonso Monsalve, Ivan Soto, Pedro Leon, Mitzi Spesny, and Andreas Busse. We would also like to acknowledge Colciencias for funding (CODIGO 11150412976) and Hospital Universitario San Vicente de Paul where the clinical work was done.




