Cohesins: Crossroad Between Cornelia de Lange Spectrum and Cancer Predisposition
Funding: This work was supported by European Cooperation in Science and Technology (COST), Fondazione AIRC per la ricerca sul cancro ETS (IG-2018 n. 21999 and IG-2023 n. 29175 to G.C.) and University of Milan fundings (to V.M. and C.G.) and Fondazione Mariani to A.S.
ABSTRACT
The cohesin complex plays crucial roles in DNA repair, chromatid separation, and gene transcription regulation. Pathogenic variants in cohesins or dysfunctional transcriptional regulators lead to cohesinopathies, a broader group of disorders including Cornelia de Lange Spectrum (CdLSp), for which the prevalence of cancer cases remains unclear. Here, we aimed to assess the prevalence of oncological events in CdLSp and elucidate the role of cohesin variants in cancer predisposition. We developed a custom next-generation sequencing (NGS) panel targeting predisposition and pathogenic genes, which we applied on N = 120 samples of pediatric patients with acute lymphoblastic leukemia (ALL), identifying 11 out of 229 total—10 germline and 1 somatic—variants in cohesin genes. Data of N = 205 brain tumors were extracted by bioinformatic analysis of data from open-source databases carrying 19 somatic variants. In a cohort of 54 CdLSp patients, the largest cohort from a single center, with a median age of 13 years, the hypothesis of an increased prevalence of cancer in CdLSp was not confirmed. Our findings highlight a significant involvement of germline NIPBL variants in CdLSp, whereas RAD21 and STAG1/2 are predominantly found as somatic variants in neoplasms. However, a distinct genetic or molecular pattern distinguishing variants leading to CdLSp from tumors was not identified. Hence, we advocate for further investigation into the relationship between cohesin variants and cancer predisposition in a larger cohort of patients, with a longer observation time and including different types of malignancies, with more focus on epigenetic approaches.
1 Introduction
Cornelia de Lange Spectrum (CdLSp) is a rare genetic congenital disorder characterized by intellectual disability, pre- and postnatal growth failure, typical facial dysmorphisms, and upper limb anomalies. CdLSp affects approximately one child in 10,000–30,000 liveborn (Kaur et al. 2023). The clinical presentation is highly variable, ranging from mild to more severe forms, more frequently burdened by chronic medical complications such as GERD, hearing impairment, epilepsy, and communication problems (Kline et al. 2018). This syndrome belongs to the broader group of disorders of transcriptional regulation (TDR), formerly known as cohesinopathies (Izumi 2016). Underlying the syndrome are germline mutations in seven genes: NIPBL, SMC1A, SMC3, RAD21, BRD4, HDAC8, and ANKRD11, encoding for proteins involved in chromatin regulation, most commonly structural or regulatory components of the cohesin complex (Selicorni et al. 2021). The cohesin complex is an evolutionarily conserved protein complex in eukaryotes that has two main functions in cells: the canonical functions consist of DNA repair and controlling sister chromatid segregation during cell division, while the non-canonical functions involve gene transcription regulation and 3D genome organization (Kline et al. 2019).
The prevalence of cancer cases in the CdLSp population has not been well established yet. The international consensus statement on Cornelia De Lange Syndrome held in 2018 stated that there is no increased risk of cancer at a pediatric age among affected patients while highlighting the unavailability of reliable data for middle-aged and elderly individuals (Kline et al. 2018). Indeed, in the literature only two papers can be found addressing CdLSp patients in young adulthood: Kline et al. (2007) evaluated a population of 49 patients (with a range of age from 11 to 50 years and an average age of 17 years) and found a 10% prevalence of Barrett's esophagus (BE), but without any evidence of cancer cases; later, Mariani et al. (2016) conducted a study on 73 patients affected by CdLSp (with an age range from 15 to 49 years) and reported eight cases of BE, again with not a single case of malignancy.
The subject warrants further study considering that cohesin genes, causative of CdLSp, are recognized to be tumor drivers when mutated at the somatic level (Pallotta et al. 2023) in several malignant tumors, including leukemia and brain solid tumors.
Indeed, in recent decades, significant strides have been made in comprehending the genetic basis of leukemogenesis, marked by a thorough delineation of somatic structural DNA rearrangements and sequence mutations commonly disrupting the lymphoid compartment (Iacobucci and Mullighan 2017). Recent evidence underscores the role of genetic predisposition in approximately 5%–10% of pediatric cases of tumors, including leukemias, even in non-syndromic patients (Inaba and Mullighan 2020). Yet, the prevalence and range of predisposing mutations among children and adolescents remain largely unknown (Zhang et al. 2015). Interestingly, there is growing evidence of the overlap of different germline and somatic variants affecting the same genes involved in predisposition and tumor progression (Vali-Pour et al. 2022).
In particular, acute lymphoblastic leukemia (ALL) stands as the predominant form of childhood cancer (InabaHiroto et al. 2013). In about 85% of childhood ALL cases, the malignant progression primarily affects B-lineage lymphocytes, triggered by various genetic anomalies, including chromosomal translocations, numeric alterations of chromosomes, and specific genetic modifications that significantly influence prognosis, risk stratification, and treatment decisions (Inaba and Mullighan 2020; Jerchel et al. 2018).
Somatic mutations in cohesin genes, renowned for their pivotal involvement in cell cycle regulation and DNA repair mechanisms (Brooker and Berkowitz 2014; Tothova et al. 2021), have been documented in myeloid malignancies (10%–20% of acute myeloid leukemia, 50% of Down syndrome acute megakaryoblastic leukemia, 5%–15% of myelodysplastic syndrome, and 10% of myeloproliferative neoplasm) (Fisher et al. 2017; Thota et al. 2014) as well as solid tumors (De Koninck and Losada 2016; Solomon et al. 2014). More recently, somatic mutations in cohesin genes have also been documented in childhood ALL (Brady et al. 2022). Of relevance, our previous research has suggested a link between ALL and cohesins, describing the first case of a patient affected by ALL and CdLSp (Fazio et al. 2019).
Interestingly, in pediatric brain tumors, cohesin pathogenetic variants have been found in glioblastoma multiforme and medulloblastoma, a rare embryonic brain tumor localized in the hindbrain (Northcott et al. 2019). Since the cohesin complex is involved in many physiological and pathological processes, including the first stages of central nervous system development (Bettini et al. 2018), gene expression regulation during embryogenesis, and DNA signaling repair (Nishiyama 2019), cohesin genes represent excellent candidates for pediatric brain tumor studies.
Overall, the aim of the present study was to assess the prevalence of oncological events in a relevant cohort of Cornelia De Lange patients at any age of life and to assess in detail possible cohesin variants in ALL and pediatric brain tumors with the ultimate goal of untangling the link among cohesin variants, CdLSp, and cancer predisposition.
2 Methods
2.1 Patients
The sample population was composed of 54 patients affected by CdLSp recruited through referral to the outpatient clinic of Sant'Anna Hospital of Como for first genetic and/or follow-up assessments between 2018 and 2023. The oncological history was obtained by consulting the medical records and by direct interviews with caregivers. Diagnosis of CdLSp was confirmed by a molecular test in all patients. To assess the presence of statistically significant associations, analyses based on the chi-square test or Fisher's test were performed. Statistical significance was defined as p-values < 0.05.
The investigation included 120 consecutive pediatric cases of ALL enrolled in the AIEOP-BFM ALL 2009 protocol across Italian AIEOP centers. These patients received their diagnoses between May and September 2016. Among them, 11 individuals were diagnosed with T-cell ALL, 107 with B-cell ALL, and 2 cases exhibited a mixed phenotype. The median age of diagnosis for patients was 4 years (with a range from 1 to 17 years). The germline origin of identified variants was assessed by sequencing of bone marrow DNA from patients in molecular remission of disease (documented by a Minimal Residual Disease [MRD] value below 5 × 10e−4).
Furthermore, the ICGC Data Portal (Release 28, March 27, 2019) was interrogated to extrapolate data on pediatric brain cancer subject variants. In particular, the projects Pediatric Brain CAncer PBCA-US and PBCA-DE were used. The two datasets comprise pediatric male and female patients from 1 to 19 years old with no specification of the type of brain tumor diagnosed; all the details are reported in Table S1. Only cases with one variant in one specific cohesin gene were considered.
2.2 Sequencing
To investigate cohesin variants in ALL, we developed a custom next-generation sequencing (NGS) panel comprising 39 genes implicated in leukemia predisposition and pathogenesis, categorized into six classes based on their biological functions (Figure 1). This panel was designed with the Integrated DNA Technology (IDT) platform (xGen Predesigned Gene Capture Pools—accessible at https://idtdna.com/site/order/ngs), which produces high-fidelity single-strand DNA probes. The panel consists of 1520 probes, collectively targeting a region spanning 141 kb.

Target-Capture NGS analysis was conducted on DNA samples extracted from bone marrow at the onset of the disease, utilizing the Nextera Flex for Enrichment protocol by Illumina (#1000000048041 v01). The pooled libraries underwent paired-end sequencing (2 × 150) on a flow cell equipped with v2.5 chemistry using the Nextseq550 instrument by Illumina. FASTQ files were generated via the Local Run Manager software.
2.3 Data Analysis
Bioinformatic analysis was carried out by the Sophia DDM software on the generated FASTQ files, which have been deposited in the ArrayExpress database under accession number E-MTAB-14362. Alignment was executed against the Human Reference sequence GRCh37/Hg19. The present study reported variants in NIPBL, RAD21, STAG1, STAG2, SMC1A, SMC3, and HDAC8 genes. Variants were filtered based on criteria including a variant fraction (VF) greater than 5% and coverage of at least ×500. The variant allelic fraction (VAF) threshold in the population was set as less than 1%. We considered variants classified as certainly pathogenic, potentially pathogenic, variants of unknown significance (VUS), and novel exonic non-synonymous variants. Variants classified as benign/likely benign in all prediction databases were excluded from the results.
Variants were interpreted according to the guidelines of the American College of Medical Genetics and Cytogenetics (Richards et al. 2015), which identifies five categories based on criteria using typical types of variant evidence (e.g., population data, computational data, functional data, segregation data). The interpretation of genetic testing has been performed in a Laboratory accredited for Medical Genetic diagnostics by clinical molecular geneticists. We consulted several common prediction databases including ClinVar, Clinical Genome, Varsome, InterVar, and COSMIC (last review July 17, 2024). Variants classified as benign/likely benign in all prediction databases were excluded from the results.
Concerning the pediatric solid tumors, variants in the coding sequence and 5′ and 3′ UTR of NIPBL, RAD21, STAG1, STAG2, SMC1A, and HDAC8 genes were evaluated. Specifically, overall variants were analyzed with Varsome (Kopanos et al. 2019) v11.17.0 June 2024 Franklin (https://franklin.genoox.com—Franklin by Genoox), and CancerVar v1.1.2 (https://cancervar.wglab.org/index.php), online available bioinformatic software tools for the interpretation of pathogenicity.
We considered variants classified as certainly pathogenic, potentially pathogenic, VUS, and novel exonic non-synonymous variants. Variants classified as benign/likely benign in all prediction databases were excluded from the results.
3 Results
To investigate the possible association/correlation between cohesin variants, CdLSp, and cancer predisposition, we analyzed three cohorts of patients affected by CdLSp or ALL, or brain tumors. Variant details are extensively reported in Tables S1–S3, and the results obtained for each group are presented below.
3.1 CdLSp Study
Fifty-four patients were evaluated. The male-to-female ratio was 0.7:1. The mean age was 13 years 7 months, with a range of 1–50 years. Subdividing the patients by age group resulted in: 35 patients (79%) aged 1–14 years, 22 patients (50%) aged 15–30 years, and 2 patients (4%) aged 31–50 years. The total number of patient-years amounted to 647. All patients were Caucasian from different Italian regions.
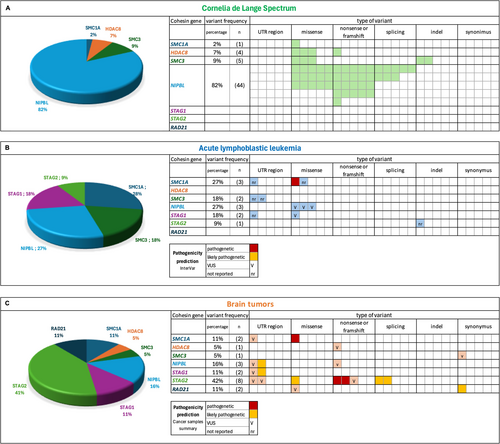
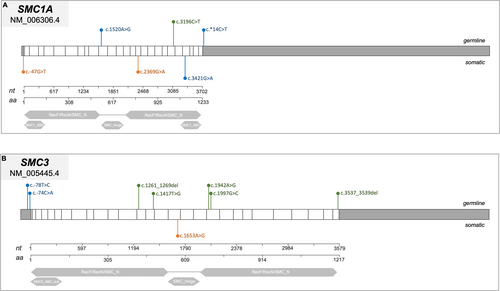
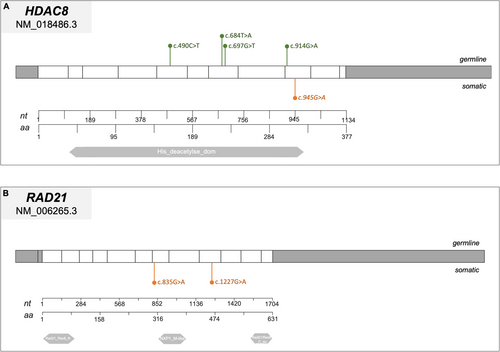
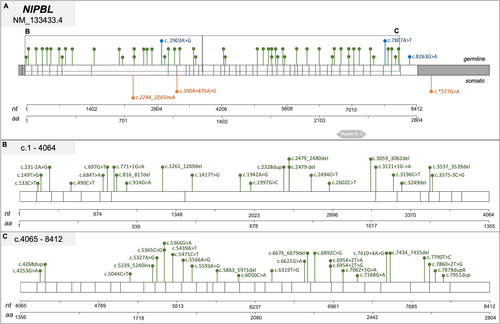
While 44 patients (82%) presented a NIPBL gene mutation, 5 (9%) had a SMC3 mutation, 4 (7%) a HDAC8 mutation, 1 (2%) a SMC1A mutation, and 1 (2%) an ANKRD11 mutation (c.1459G>T). Regarding the mutational types, 21 (47%) result in truncating protein (11 were nonsense mutations and 10 were frameshift mutations), 11 (25%) were missense mutations, and 8 (18%) were splice site mutations (Figures 2A and 3-6). Three CdLSp patients (5.5%) had a germinal mosaicism condition (Table S1).
The oncological history was largely negative, except for a significant finding of Barrett's esophagus (classified as a precancerous condition), of which six cases were identified, giving a prevalence of 9.7%. Out of 62 patients, only one received a diagnosis of a benign tumor: a 28-year-old male diagnosed with a pituitary adenoma at the age of 12. One case remained doubtful: at the biopsy performed by esophagogastroduodenoscopy, micronodules of smooth muscle tissue compatible with muscularis mucosae were identified, but the possibility of esophageal leiomyomas could not be excluded. We did not find any cases of malignant tumors (Table 1). The occurrence of all the events in patients carrying mutations in NIPBL reflects the distribution of this gene mutation in the sample population.
| Tumor histotype | Classification | Age at diagnosis (years) | Gender | CdLSp mutation |
|---|---|---|---|---|
| Pituitary adenoma | Benign | 12 | M | NIPBL (c.6319A>G) |
| Barrett's esophagus | Precancerous | 19 | F | NIPBL (c.3059_3062delAGAG) |
| Barrett's esophagus | Precancerous | 18 | F | NIPBL (not available) |
| Barrett's esophagus | Precancerous | 14 | M | NIPBL (c.5471C>T) |
| Barrett's esophagus | Precancerous | 24 | M | NIPBL (not available) |
| Barrett's esophagus | Precancerous | 34 | F | NIPBL (c.5566A>G) |
| Barrett's esophagus | Precancerous | 16 | M | NIPBL (not available) |
| Esophageal leiomyoma (uncertain) | Benign | 5 | M | NIPBL (c.771 + 1GA) |
3.2 ALL Study
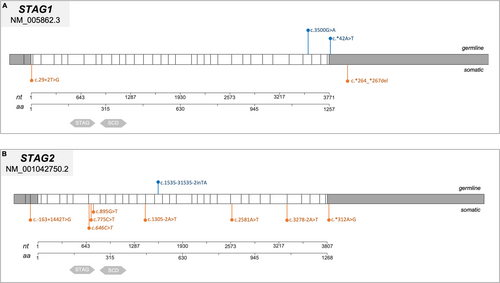
Cohesin gene variants were found in 10 out of 120 cases (8%) and these patients were carriers of 11 cohesin variants (5%, 11/229). Variants in cohesin genes were identified in 5% of cases (11 out of 229 total variants). Specifically, STAG1 and SMC3 each carried two variants, NIPBL and SMC1A had three variants each, while only one variant was found in STAG2. Notably, no pathogenetic variants were detected in HDAC8 or RAD21 (Figures 2B and 3-6).
NGS sequencing of remission-phase bone marrow samples from 10 patients with cohesin gene variants revealed that 10 out of 11 (91%) were present during the disease-negative phase, confirming their germline origin. The remaining variant in SMC1A was somatic.
We mapped the variants across the cohesin genes to define their specific localization within the gene and protein structure (Figures 3-6). Of the 11 aberrations, 7 were exonic, while 4 were located at the 5′–3′ UTR regulatory regions. Pathogenicity of the 10 germline variants was assessed using multiple online prediction tools, detailed results are summarized in Table S2.
In particular, only two variants in NIPBL were reported by both ClinVar and InterVar, while five were present in only one of the two tools. On the other hand, Varsome classified the majority of variants (7/11) as benign or likely benign, one as a variant of unknown significance (VUS), and three variants were unreported. Franklin was the only tool that made predictions for all variants, categorizing most (8/11) as VUS. Overall, the combined results from all tools did not provide clear evidence of pathogenicity for any variant, leading us to classify them all as VUS.
3.3 Pediatric Brain Tumor Study
Exploiting PBCA-US (290 patients) and PBCA-DE (541 patients) projects on the ICGC Data Portal, we identified 205 subjects carrying somatic variants in cohesin genes. Among them, 19 bearing a single variant in one cohesin gene inquired in coding or 5′/3′ untranslated regions. In particular, the 19 variants found were distributed as follows (Figures 2-5): STAG2 8/19 (42%), SMC1A 2/19 (11%), NIPBL 3/19 (16%), HDAC8 1/19 (5%), STAG1 2/19 (11%), RAD21 2/19 (11%), SMC3 1/19 (5%) (Figure 2C). Most variants were single-base substitutions and their positioning along the gene is shown in Figures 3-6. We analyzed the 19 variants using different available online platforms for pathogenicity prediction, and the results are summarized in Figure 2C. In particular, Varsome predicted the majority of variants to be benign except for two, which were classified as VUS (Table S3). Among the associated databases considered by Varsome, we also investigated the Cancer samples summary that aggregates different cancer databases, giving a more focused prediction on cancer variants than the overall somatic Varsome output. As shown in Table S3, the Cancer sample summary predicted most of the variants, with a more pathogenetic score compared to Varsome.
Franklin database identified most variants as VUS and 5 out of 19, exclusively in the STAG2 gene, as pathogenic.
Last, we considered the CancerVar database for the interpretation of somatic variants identified in pediatric brain tumor patients. For 8 out of 19 variants, CancerVar predicted the pathogenicity as VUS or likely pathogenic. Notably, taking together all software results, we identified exclusively two variants in STAG2 with a likely pathogenic/pathogenic prediction (Table S3).
4 Discussion
CdLSp is a genetic disease caused by germline mutations in the cohesin genes that affect many organ systems. A recent systematic review suggested that CdLSp could be a cancer-predisposing condition due to the common knowledge that genetic syndromes characterized by mutations in genes codifying for proteins having the function of DNA repair can determine a predisposition to the development of neoplastic events, together with the discovery of mutations in cohesin genes in certain tumors (Pallotta et al. 2023).
To clarify the occurrence of cancer in CdLSp, we surveyed, for the first time, 54 patients with a molecular diagnosis of CdLSp belonging to different age groups, and we did not identify any case of malignancy.
We observed, though, an increased prevalence of Barrett's esophagus, that is closely related to chronic GERD, which De Lange patients are known to suffer from (Kline et al. 2007; Macchini et al. 2010). The increased risk of BE at a young age was already known in patients with CdLSp (Kline et al. 2007; Luzzani et al. 2003; Macchini et al. 2010; Mariani et al. 2016); this study further confirms this observation, having identified six cases of BE (with a prevalence corresponding to 9.7%) at an average age of 20 years 8 months. Hence, the hypothesis that CdLSp may be a cancer-predisposing condition is not supported by the present study.
However, given the increasing evidence of an association between cohesin genes and cancer, we investigated cohesin variants in two cancer cohorts, that is, pediatric ALL and brain solid tumors, for which a hypothesis of cohesin genes involvement was so far more plausible, also considering that they are the most frequent types of cancer in childhood. In both population studies, we found variants in the cohesin complex genes, and in the ALL study, we were able to ascertain the germline origin in 10/11 identified variants, which are classified as VUS according to the Franklin algorithm. By definition, VUS have an unclear phenotypic impact. However, in a multistep model for tumorigenesis, such variants could promote a condition favorable to acquire additional hits and ultimately lead to a clinically overt disease (i.e., ALL). The identified variants do not have sufficient pathogenicity to generate a cohesinopathy and, thus, certainly do not translate into a syndromic manifestation, as, on the contrary, variants with a strong pathogenic prediction are supposed to. They could thus be associated with an increased risk of leukemia incidence, as previously demonstrated in the case of germline variants in the STAG1 gene (Saitta et al. 2022). Further, in our ALL cohort, the majority of variants affecting cohesins were of germline origin. Only one patient exhibited a somatic second hit in SMC1A alongside a germline variant. This scenario of potential double-hit genetic events exemplifies how a germline variant may promote genomic instability, thereby predisposing to additional somatic mutations that contribute to disease onset, as we have recently hypothesized in the case of ALL in a Cornelia de Lange patient (Fazio et al. 2019).
Considering these observations, we further compared variants among the three study populations. Overall, as expected, we observed a major involvement of NIPBL in CdLSp, whereas RAD21 and STAG1/STAG2 were mainly found in cancer subjects. However, when analyzing the genetic position and/or affected domains as well as the type of variants, we did not observe a clear pattern in CdLSp compared to either ALL or brain tumor samples. Hence, despite the intrinsic limit of the study due to the small number of cases, we were not able to identify the molecular mechanisms underlying CdLSp vs. cancer path. To tackle this scientific enigma, we believe that the next step would be either (i) enlarging the study cohort and the observation time to include also patients with older age; (ii) extending the functional analysis of germline cohesin variants to ascertain their contribution to DNA repair and/or gene regulation, and (iii) studying the episignature in these—or similar—three cohorts. Indeed, there is mounting evidence that CdLSp is clearly associated with a specific episignature, irresponsive to the genetic causes (Peng et al. 2024), shifting the riddle from genetics to epigenetics. Lastly, many congenital syndromes are caused by genes that play a role also in oncogenesis (e.g., Rubinstein-Taybi OMIM# 180849 or Noonan OMIM# 163950), but only in some cases do the syndrome-causing mutations result in an increased risk for cancer (Roberts et al. 2013; Roth et al. 2024). Hence, the necessity of deepening such associations with a wider lens, that is epigenetics or cellular cascades.
In conclusion, according to our data, in this still limited subset of unselected CdLSp patients, with a median age of 13 years, the hypothesis of an increased prevalence of cancer in CdLSp syndromic patients does not seem to be confirmed. Because of the relevance of this information, it is of paramount importance to increase such data collection and observation time in order to obtain a better assessment of patients' clinical counseling.
Author Contributions
Laura Rigotti: data collection, analysis, writing, and editing. Stefano Rebellato: data collection, analysis, writing, and editing. Antonella Lettieri: data collection, analysis, writing, and editing. Grazia Fazio: data collection, analysis, writing, and editing. Silvia Castiglioni: images and data analysis. Milena Mariani: data collection and analysis. Simona Totaro: data collection and analysis. Claudia Saitta: data collection and analysis. Cristina Gervasini: reviewing and editing. Valentina Massa: conceptualization, writing, reviewing, editing, and funding. Giovanni Cazzaniga: conceptualization, writing, reviewing, editing, and funding. Angelo Selicorni: conceptualization, writing, reviewing, editing, and funding.
Acknowledgments
We thank all the funding bodies, in particular this work was partly supported by the European Cooperation in Science and Technology (COST); by grants from the Fondazione AIRC per la ricerca sul cancro ETS (IG-2018 n. 21999 and IG-2023 n. 29175 to G.C.) and University of Milan fundings (to V.M. and C.G.) and Fondazione Mariani to A.S. S. Rebellato is a fellow of the University of Milano-Bicocca, Milan, Doctoral Program in Molecular and Translational Medicine (DIMET). S. Totaro is a fellow of the University of Milan, Milan, Doctoral Program in Translational Medicine. S. Castiglioni is a fellow of the Medical Genetics School of the University of Milan, Milan. A. Lettieri is a fellow of Fondazione Veronesi. We thank all the funding bodies, in particular this work was partly supported by the EU-COST
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.




