A Novel Heterozygous c.1024A>G Variant in BMPR1B Causes Either Isolated Brachydactyly Type A4 With Variable Expressivity or Incomplete Type A4 Overlapping Type D in a Chinese Han Pedigree
Funding: This work was supported by the Southwest Medical University (Grant No. 00040164 and 202310632061) and the Laboratory of Xinjiang Endemic and Ethnic Diseases at Shihezi University, Ministry of Education (Grant No. KF202401).
Xinyi Yang and Xiaqing Wu contributed equally to this work.
ABSTRACT
BDA4 and BDD are rare autosomal dominant conditions characterized by distinct hand/foot malformations, including middle phalangeal shortening in the second and fifth digits and short, broad thumb terminal phalanges. While variations in BMPR1B have been implicated in the pathogenesis of BDA1 and BDA2, the genetic basis underlying BDA4 and BDD remains unclear. Clinical and radiographic phenotyping were performed to assess and diagnose the affected pedigree. Whole-exome sequencing and Sanger sequencing were employed to identify and validate the genetic variation. Bioinformatics analyses were conducted to evaluate the potential pathogenicity of the variant. Functional validation was carried out by assessing SMAD4 localization in BMP4-stimulated 293T transfectants. We present the first report of a rare Chinese Han pedigree exhibiting two distinct phenotypes: isolated BDA4 and incomplete BDA4 overlapping BDD, which were observed across two branches. All affected individuals harbored a novel heterozygous c.1024A>G (p.K342E) variant in BMPR1B, with bioinformatics analyses suggesting its pathogenic potential. Structural analyses indicated a conformational change within the kinase domain. Functional assays revealed a marked reduction in nuclear SMAD4 accumulation in transfectants expressing the mutant BMPR1B compared to the wild-type counterpart. This study provides the first evidence implicating BMPR1B as a pathogenic gene for both isolated BDA4 and incomplete BDA4 with BDD overlap. The BMPR1B c.1024A>G (p.K342E) variant disrupts kinase activity and impairs SMAD1/5/8 phosphorylation, which in turn suppresses downstream IHH expression and interferes with BMP-mediated skeletal patterning. We propose that the variant, in combination with genetic background and environmental factors, leads to the observed variable expressivity in this pedigree. Our findings expand the mutational spectrum of brachydactyly and underscore BMPR1B as a candidate gene for further investigation in brachydactyly pathogenesis.
1 Introduction
Brachydactyly (BD) manifests either as an isolated malformation or as a symptom of syndromic disorders of limb dysplasias. Patients with BD claim to state fine motor inconvenience and poor shape in digits or toes. As a component of complex malformation syndromes, BD is observed in Robinow syndrome (OMIM: 268310), Du Pan syndrome (OMIM: 228900), Temtamy preaxial brachydactyly syndrome (OMIM: 605282) and other BD-related syndromes. As an isolated disease, BD represents an autosomal dominant hand malformation characterized by bone hypoplasia with variable patterns of phalangeal or metacarpal shortening/absence, potentially accompanied by foot deformities (Temtamy and Aglan 2008; Temtamy and McKusick 1978). BD was initially classified by Bell based on affected digit patterns and further elaborated by Temtamy and Aglan (2008). Current classification delineates isolated BD into types A–E (BDA–BDE), where BDA is further subdivided into subtypes 1–4 (BDA1–BDA4). Clinically, BDA3 and BDD demonstrate higher prevalence compared to other subtypes (Temtamy and McKusick 1978).
BDA4 (OMIM: 112800), alternatively termed Temtamy type brachydactyly or brachymesophalangy II and V, primarily affects the second and fifth digits through middle phalangeal shortening. When involving the fourth digits, characteristic radial clinodactyly of the distal phalanges occurs due to abnormally shaped middle phalanges. BDA4 patients typically exhibit toe malformation and dysostoses, predominantly affecting the lateral four toes. Some BDA4 patients present ulnar deviation of the second digits and radial deviation of the fifth digits (Temtamy and Aglan 2008; Temtamy and McKusick 1978). BDD (OMIM: 113200), colloquially termed Stub thumb, manifests as a distinctively short and broad terminal thumb phalanges, generally with no consistent skeletal associations (Johnson et al. 2003; Robin et al. 1999). BDD appears bilaterally in approximately 75% of cases, often with symmetrical involvement (Robin et al. 1999; Gray and Hurt 1984). Accumulating evidence implicates HOXD13 as the principal genetic determinant of BDD, with specific variants demonstrating phenotypic overlap between BDD and BDE (Johnson et al. 2003; Robin et al. 1999; Thomsen 1928). Notably, HOXD13 emerges as a strong candidate for syndromic BDA4, given its association with synpolydactyly-brachydactyly syndrome displaying a BDA4-like phenotype (Temtamy and Aglan 2008; Zhao et al. 2007; Zhang et al. 2020). Nevertheless, the pathogenic genes for isolated BDA4 remain undefined.
The bone morphogenetic protein (BMP) signaling pathway plays a pivotal role in phalangeal development, comprising ligands BMP1-15, growth differentiation factor 5 (GDF5), BMP serine–threonine kinase receptors type 1A (BMPR1A), type 1B (BMPR1B), type 2 (BMPR2), and their antagonist noggin (NOG) (Figure 1A). BMP ligands bind to heteromeric receptor complexes composed of BMPR1A, BMPR1B, and BMPR2 at the cell membranes (Horbelt et al. 2012). The activated BMPR1B phosphorylates downstream SMAD1/5/8, which then complexes with co-factor SMAD4 and translocates to the nucleolus. The transcriptional complex ultimately activates Indiana Hedgehog (IHH) expression via interaction with transcription factor inhibitor of DNA binding (ID), thereby regulating digit morphogenesis (Seki and Hata 2004; Lyu et al. 2019). BMPR1B, a 502-amino-acid transmembrane protein, contains three main functional domains, namely: an extracellular ligand-binding domain, a single transmembrane domain, and an intracellular protein kinase domain. As an integral component of the BMP signaling pathway, BMPR1B variants have been implicated across a spectrum of limb dysplasias (Table 1), particularly within its kinase domain (Figure 1B) (Racacho et al. 2015). However, current evidence linking BMPR1B variants to BD remains limited. Reported cases include: heterozygous p.K325N missense variant and heterozygous c.447-1G>A splice variant in BMPR1B in two unrelated children exhibiting BDA1 (Racacho et al. 2015), heterozygous variants of p.I200K and p.R486W in BMPR1B in two unrelated German families possessing BDA2 (Lehmann et al. 2003), and heterozygous missense p.R486Q/L variants in BMPR1B causing BDA2 (Lehmann et al. 2006; Badura-Stronka et al. 2015). Notably, no BMPR1B variants have been conclusively associated with BDA4 or BDD phenotypes to date.
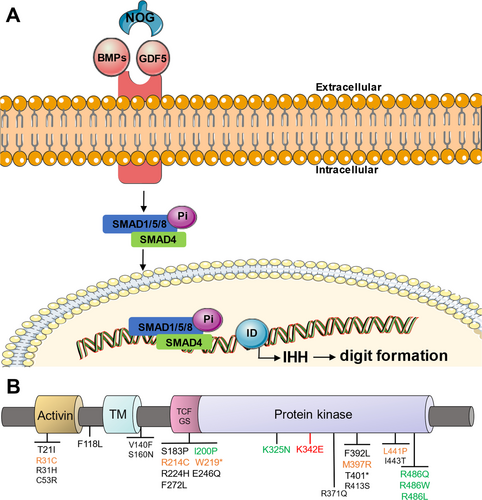
| Encoded Substitution | Phenotype | Reference | |
|---|---|---|---|
| 1 | T21I | Pulmonary arterial hypertension? | (Yang et al. 2018) |
| 2 | R31H/C | Chondrodysplasia, du Pan type, and Iron overload? | (Stange et al. 2015; Schaefer et al. 2015) |
| 3 | C53R | Acromesomelic chondrodysplasia, Grebe type | (Graul-Neumann et al. 2014) |
| 4 | F118L | Pulmonary arterial hypertension, idiopathic? | (Ban et al. 2018) |
| 5 | V140F | Pulmonary arterial hypertension, idiopathic? | (Ban et al. 2018) |
| 6 | S160N | Pulmonary arterial hypertension | (Chida et al. 2012) |
| 7 | S183P | Pulmonary arterial hypertension | (Kim et al. 2018) |
| 8 | I200P | Brachydactyly type A2 | (Lehmann et al. 2003) |
| 9 | R214C | Complex digit malformations | (Yıldırım et al. 2018) |
| 10 | W219* | Acromesomelic chondrodysplasia, Grebe type | (Graul-Neumann et al. 2014) |
| 11 | R224H | Primary ovarian insufficiency? | (Patiño et al. 2017) |
| 12 | E246Q | Pulmonary arterial hypertension, idiopathic? | (Ban et al. 2018) |
| 13 | F272L | Primary Ovarian Insufficiency | (Renault et al. 2020) |
| 14 | K325N | Brachydactyly type A1 | (Racacho et al. 2015) |
| 15 | R371Q | Pulmonary arterial hypertension, idiopathic | (Ban et al. 2018) |
| 16 | F392L | Pulmonary arterial hypertension | (Chida et al. 2012) |
| 17 | M397R | Acromesomelic dysplasia, Hunter-Thompson type | (Ullah et al. 2018) |
| 18 | T401* | AMD Grebe type | (Lhousni et al. 2023) |
| 19 | R413S | Pulmonary arterial hypertension? | (Yang et al. 2018) |
| 20 | L441P | Du Pan syndrome | (Turgut et al. 2023) |
| 21 | I443T | Pulmonary arterial hypertension, early-onset? | (Zhu et al. 2018) |
| 22 | R486W/Q/L | Brachydactyly, Brachydactyly type A2 | (Lehmann et al. 2003; Lehmann et al. 2006; Badura-Stronka et al. 2015) |
- * Nonsense mutation.
In this study, we report a rare Chinese Han pedigree, presenting the first documented occurrence of two distinct phenotypes: isolated BDA4 and incomplete BDA4 overlapping BDD, simultaneously observed across two branches. To explore the etiology of this BD pedigree, Whole-exome sequencing and Sanger sequencing were performed, identifying a novel heterozygous c.1024A>G (p.K342E) variant in BMPR1B. Comprehensive bioinformatics analyses, coupled with functional assays, were conducted to validate its pathogenicity role within this BD pedigree.
2 Materials and Methods
2.1 Editorial Policies and Ethical Considerations
Informed consent was obtained from all participating subjects. The research protocol was conducted in accordance with the Declaration of Helsinki guidelines and approved by the Ethics Committee of the Affiliated hospital of Southwest Medical University (Approval No. KY2023410).
2.2 Patient Collection
Clinical examinations of hands and feet were conducted on all family individuals within this Chinese Han pedigree. All family individuals, who had no consanguineous marriage, exhibited normal stature and intellect. Except for isolated BD, no deafness, characteristic facial features, hypoplastic genitalia, or other skeletal abnormalities/syndromes were observed in this pedigree.
2.3 Photographs and X-Ray Examination
Clinical photographs of hands and feet were taken using smartphones. Radiographs of hands and feet were obtained through Digital Radiography.
2.4 Gene Sequencing
Genomic DNA was extracted from blood samples of family individuals. Whole-exome sequencing of the proband and her aunt were performed at MyGenostics Sequencing Service (Beijing, China). The whole exomes in affected individuals, along with adjacent intron regions (50 bp), were captured via the GenCap whole-exome capture kit, amplified, and high-throughput sequenced using the DNBSEQ-T7 to prepare the DNA library. Then the sequencing data were mapped to the UCSC hg19 human reference genome through the parameter BWA in Sentieon software for variation detection. Sanger sequencing was also conducted in this pedigree to verify their variation results and whether other unaffected individuals carried the same suspected variant.
2.5 Bioinformatics Analyses
Bioinformatics analyses were conducted by mapping the sequencing data to multiple databases, including OMIM, Clinvar, dbSNP, and HGMD. The variants of SNV were identified using the parameter driver of Sentieon software. Based on the Standards and Guidelines for the Interpretation of Sequence Variants of the American College of Medical Genetics and Genomics (ACMG), variation pathogenicity was assessed. SIFT, MutationTaster, GERP+, and REVEL were also employed to predict the variation pathogenicity. Clustal Omega was utilized for the conservation analyses of the variant sites across different species. In addition, I-TASSER (Yang and Zhang 2015; Zheng et al. 2021; Zhou et al. 2022) was applied to predict the spatial conformation of the protein, changes in binding ligand types, binding sites, enzyme commission (EC) number, and active sites. Pymol was used to analyze the 3D structural difference between the wild-type and mutant protein.
2.6 Plasmid Construction
Wild-type plasmid pCMV-SPORT6-BMPR1B (Human) was obtained from MiaoLingBio (Wuhan, China). The QuickMutation Plus Site-Directed Mutagenesis Kit from Beyotime Biotechnology (Shanghai, China) was used to prepare the mutant plasmid pCMV-SPORT6-BMPR1B-K342E. All vectors were verified via sequencing analyses.
2.7 Cell Culture, Transfection and Protein Localization Analyses
Human embryonic kidney epithelial cells (HKC) 293T was cultured through standard methods. Wild-type and mutant BMPR1B plasmids were transfected into HKC 293T cells using Lipo293 Plus Transfection Reagent form Beyotime Biotechnology. After 24 h of transfection, exogenous BMP4 ligand from MedChemExpress (Shanghai, China) was added to the culture medium for 48 h. The action concentration of BMP4 was 10 ng/mL. After 72 h of transfection, SMAD4 Rabbit Monoclonal Antibody labeled with green fluorescence from Beyotime Biotechnology was added, and fluorescence intensity was detected. The crawling cell coverslips were washed 3 times with phosphate-buffered saline (PBS) on the culture plate, then fixed with 4% paraformaldehyde for 15 min, washed 3 times with PBS again, and mounted with medium containing 4,6-Diamidino-2-phenylindole dihydrochloride (DAPI). The coverslips were observed, and images were acquired under an Olympus BX63 fluorescence microscope.
3 Results
3.1 Patients Report and Radiodiagnosis
The pedigree analyses revealed five affected individuals (II2, II3, III2, III3, and III4) and four unaffected individuals (I1, II1, II4, and III1) among three generations. Phenotypic assessment of the proband's grandma (I2) could not be performed due to deceased status (Figure 2A). The 21-year-old Han Chinese female proband (III3) exhibited bilateral symmetrical shortening of the middle phalanges in the second and fifth digits, radial deviation of the fourth digits, and middle-distal phalangeal synostosis affecting the fourth and fifth toes, accompanied by fifth toenail dysplasia (Figure 2J–J5,K–K5,L–L5). These manifestations were consistent with typical isolated BDA4. The proband's 53-year-old mother (II3) demonstrated bilateral mild fifth-digit middle phalangeal shortening, with asymmetric radial deviation (right > left) of the fourth digits, combined with fifth toes middle-distal phalangeal synostosis and toenail deformity (Figure 2H–H5,I–I5). The proband's 16-year-old sister (III4) presented bilateral hand clinodactyly characterized by ulnar deviation of the second digits (left > right) and radial deviation of the fourth and fifth digits (right > left), with only mild fifth-digit middle phalangeal shortening. III4 presented identical phalangeal synostosis patterns to those observed in II3, though lacking toenail deformity (Figure 2M–M5,N–N5). Accordingly, both II3 and III4 were classified as incomplete BDA4. However, the proband's aunt (II2) manifested distinct phenotypic features, including bilateral short and broad thumb distal phalanges, diagnostic of BDD. Additional findings included fifth-digit middle phalangeal shortening and fourth-digit radial deviation, without pedal involvement, suggesting incomplete BDA4 presentation (Figure 2F–F5,G–G5). These phenotypic patterns were replicated in the proband's elder cousin (III2). No radiographic evidence of brachydactyly or other skeletal abnormalities was detected in proband's father (II4) (Figure 2D–D5,E–E5), her uncle (II1) (Figure 2B–B5,C–C5) and her younger cousin (III1).
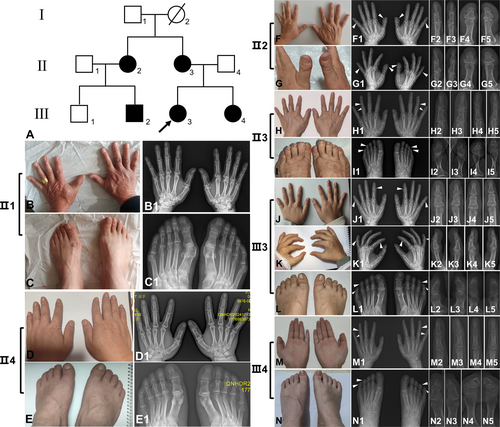
3.2 Whole-Exome Sequencing and Sanger Sequencing
Peripheral blood leukocytes were obtained from II2-4 and III1-4. Whole-exome sequencing was conducted separately on II2 and III3. Among the sequencing results, high-frequency polymorphisms and variants unrelated to skeletal development pathways were excluded. We identified a heterozygous missense c.1024A>G (p.K342E) variant in BMPR1B (NM_001203.3; exon 10) in II2 and III3, causing an amino acid substitution from K to E at position aa-108. Subsequent Sanger sequencing confirmed this variant in affected individuals II3, III2, and III4, while being absent in unaffected individuals II4 and III1 (Figure 3A–D).
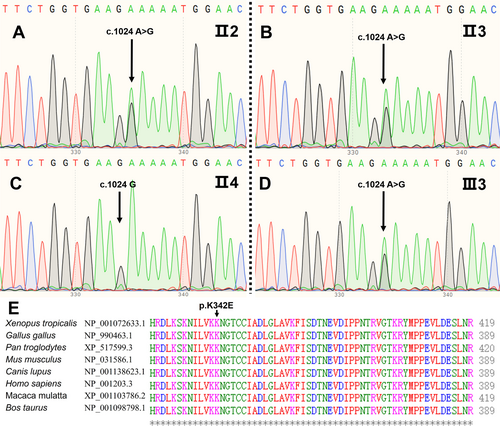
3.3 Bioinformatics Analyses
The c.1024A>G (p.K342E) variant in BMPR1B was identified as a single nucleotide variation (SNV) (ID: rs748524936) via dbSNP. Population frequency analysis using gnomAD v4 revealed an allele frequency of 0.0001114 in East Asian and 0.000008055 in the total population. This variant has not been cataloged in the Human Genome Mutation Database (HGMD) and its functional implications remain uncharacterized. SIFT predicted it to be damaging (0.022); MutationTaster analysis deemed it disease-causing (1); GERP+ predicted it conserved (6.06); REVEL predicted it LB (0.242). Clustal Omega analysis revealed absolute conservation of BMRP1B locus 342 aa across eight vertebrate species: Xenopus tropicalis , Gallus gallus , Pan troglodytes , Mus musculus , Canis lupus , Homo sapiens , Macaca mulatta , and Bos taurus (Figure 3E).
In addition, according to I-TASSER prediction results of BMPR1B conformations, Pymol was performed to construct wild-type and BMPR1BK342E variant 3D models. Comparative analyses revealed significant conformational reorganization in the mutant BMPR1B (Figure 4A–C). To elaborate, before the replacement, wild-type protein displayed a β-sheet at 338–340 aa and a reverse β-sheet at 346–347 aa. After the replacement, the BMPR1BK342E variant exhibited an extended β-sheet including 338–341 aa along with partial 342 aa, and a reverse β-sheet at 345–347 aa (Figure 4A′–C′). Subsequent hydrogen-bonding analyses identified critical perturbations at locus 342 aa. Before the replacement, an intramolecular hydrogen bond was formed between the amino hydrogen of N 284 on the amide bond and the carbonyl oxygen of N 284, while the bond length was 2.2 Å. Another hydrogen bond was formed between the carboxyl oxygen without hydrogen ions of E 283 and the protonated amino hydrogen of K 342, while the bond length was 1.8 Å (Figure 4D). After the replacement, both the hydrogen bonds mentioned above disappeared in the BMPR1BK342E variant. An intramolecular hydrogen bond was formed between the amino hydrogen of E 283 on the amide bond and the carboxyl oxygen without hydrogen ions of E 283, and the bond length was 2.0 Å. The second hydrogen bond was formed between the carboxyl oxygen without hydrogen ions of E 283 and the protonated amino hydrogen of K 227, and the bond length was 2.3 Å. The third hydrogen bond was formed between the carbonyl oxygen of E 283 and the protonated amino hydrogen of K 227, and the bond length was 1.9 Å (Figure 4E). Besides, the binding ligand types, the binding sites (Table S1), the Enzyme Commission (EC) numbers, and the active sites (Table S2) showed significant changes after the variation.
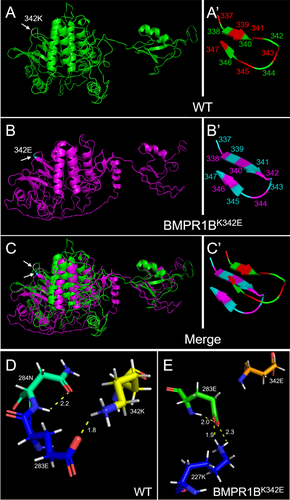
Applying ACMG criteria, the variant met two moderate-strength evidences and three supporting evidences, which are: (1) located within a mutational hotspot or a key functional domain known to be free of benign variants (PM1); (2) ultra-rare frequency in the general population in the ExAC database (PM2); (3) complete co-segregation with phenotypes across three generations (PP1); (4) the predicted destructive effect of the variant on protein function (PP3); (5) phenotypic support for the gene variant (PP4). Based on these evidences, the variant was assessed to be suspected pathogenic.
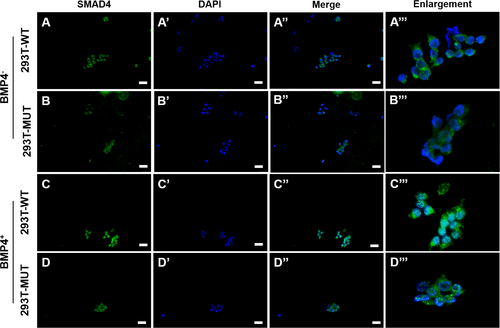
3.4 Subcellular Localization Analyses of SMAD4
In unstimulated conditions without exogenous BMP4 ligand addition, both wild-type and p.K342E mutant BMPR1B-transfected 293T cells exhibited predominant cytoplasmic SMAD4 localization, with minimal nuclear accumulation (Figure 5A–A‴,B–B‴). Following exogenous BMP4 ligand stimulation, wild-type transfectants demonstrated robust nuclear SMAD4 enrichment with residual cytoplasmic retention (Figure 5C–C‴); conversely, mutant transfectants maintained cytoplasmic SMAD4 sequestration, indicating significantly reduced nuclear translocation efficiency after the BMPR1B variant (Figure 5D–D‴).
4 Discussion
The present report describes the BD pedigree with a remarkable range of different phenotypes, where both isolated BDA4 and incomplete BDA4 overlapping BDD were identified across two branches within a single pedigree. In the proband's branch, III3 was initially designated as BDA4 based on typical middle-phalangeal shortening and deviation patterns. Comprehensively considering atypical presentation in II3 and III4, both them were categorized as incomplete BDA4 due to attenuated digital manifestations. Notably, in her aunt's branch, II2 and III2 exhibited typical BDD features, co-occurring with fourth/fifth-digit brachydactyly and clinodactyly, culminating in a novel incomplete BDA4 overlapping BDD diagnosis. By comparatively analyzing, II2 and III3 share equivalent fourth/fifth-digit anomalies despite divergent thumb morphologies, demonstrating phenotypic convergence between the two branches. A similar phenomenon has been reported that two unrelated sporadic families carrying the same BMPR1B p.R486Q variant respectively presented BDA2-syndactyly syndrome and isolated BDC (Lehmann et al. 2006), providing evidence of remarkably different BD phenotypes occurring within a pedigree simultaneously.
To investigate the observed phenotypic divergence, Whole-exome sequencing was performed on II2 and III3 separately, trying to identify multiple candidate pathogenic variants. Sanger sequencing validation confirmed a shared novel BMPR1B c.1024A>G (p.K342E) variant across all affected individuals, which has not been previously described in the literature and has not been reported in the Human Genome Mutation Database so far. Bioinformatics analyses conducted in our study yielded compelling evidence that the variant was pathogenic for BD. Intriguingly, the identical variant manifested as either isolated BDA4 with variable expressivity or incomplete BDA4 overlapping BDD, which is hard to explain by current theoretical knowledge.
As far as we have concerned, three possible mechanisms may underlie this genotype–phenotype dissociation. Firstly, our study has demonstrated significant conformational reorganization of the BMPR1BK342E variant. Previous studies identified the p.K325N and p.R486Q/W/L variants in BMPR1B for BD (Table 1) (Racacho et al. 2015; Lehmann et al. 2003; Lehmann et al. 2006; Badura-Stronka et al. 2015), both located within the BMPR1B kinase domain (Figure 1B). Lemuel Racachoa et al. reported a BMPR1B c.975A>C (p.K325N) variant identified in individuals displaying BDA1. This pathogenic variant lies in the BMPR1B kinase domain, and the expression of BMPR1BK325N led to an ~50% reduction in reporter gene activity, which acts as a dominant-negative variant (Racacho et al. 2015). Katarina Lehmann et al. found a c.1456C>T (p.R486W) variant in a highly conserved region C-terminal of the BMPR1B kinase domain. Micromass culture experiments (in vitro) showed that BMPR1BR486W in chBMPR1B affects cartilage formation in a dominant-negative manner, and overexpression tests in chicken embryos (in vivo) also showed shortening and/or absence of individual phalanges (Lehmann et al. 2003). In our research, structural modeling revealed significant conformational alterations in the BMPR1BK342E variant, which is spatially proximal to the previously characterized pathogenic BMPR1BK325N variant within the kinase domain, suggesting impaired kinase functionality as a plausible pathogenic mechanism in BD. To verify this possibility, functional experiments were conducted, demonstrating a significant reduction in nuclear SMAD4 accumulation in transfected BMPR1BK342E mutant 293T cells compared to the wild-type controls through BMP4 stimulation. These findings substantiate structural perturbation-induced BMPR1B kinase activity. In future investigations, we will strive to incorporate chondrocyte models to further assess developmental pathway dysregulation. Secondly, considering BMPR1B as a component of the BMP signaling pathway, we deem that the divergent expressions in this pedigree are associated with abnormal expression of the BMP signaling pathway. We found the substitution from K to E at position 342 aa results in ligand-binding changes in ligand types and binding sites (Table S1). Wild-type BMPR1B exhibited affinity interactions with ADP, 03Q, and GBL, while the BMPR1BK342E variant acquired neo-binding capacity for SS3, 4BM, and W2R. According to the experimental results, this ligand switching correlated with decreased SMAD1/5/8 phosphorylation, thus reducing SMAD4 complex formation, resulting in impaired nuclear translocation of the SMAD1/5/8-SMAD4 complex. Consequently, downstream IHH expression was suppressed, disrupting BMP-mediated skeletal patterning and ultimately causing BD(Figure 1A). The BMPR1B variant may also dysregulate the intricate equilibrium among the ligands, inhibitors, and receptors, indirectly interfering with IHH expression. Thirdly, we propose that remarkable phenotypic divergence emphasizes the crucial role of genetic background in combination with environmental factors interacting with the c.1024A>G variant in BMPR1B during embryonic development. A similar mechanism was previously reported in three sporadic pedigrees carrying the same variant (Savarirayan et al. 2003). Given the rarity of this BD type, our study provides promising initial findings. However, we emphasize the critical importance of independent replication studies and functional validation to confirm these results. Elucidating the precise molecular interplay will require further investigations.
5 Conclusion
This study provides the first evidence implicating BMPR1B as a pathogenic gene for both isolated BDA4 and incomplete BDA4 overlapping BDD. The novel heterozygous BMPR1B c.1024A>G (p.K342E) missense variant simultaneously induces isolated BDA4 with variable expressivity and incomplete BDA4 overlapping BDD across two branches within a Chinese Han pedigree. Mechanistically, the conformational reorganization of the BMPR1BK342E variant impaired kinase functionality and reduced SMAD1/5/8 phosphorylation efficiency, leading to dysregulation of the BMP signaling pathway. We hypothesize that the genetic background, in conjunction with environmental factors during embryonic development, interacts with the c.1024A>G variant in BMPR1B to produce the observed phenotypic variability within this BD pedigree. These findings broaden the mutational spectrum of BD and establish BMPR1B as a critical gene for further investigation in BD pathogenesis, offering potential for improved molecular diagnostics upon validation in larger cohorts.
Author Contributions
Xinyi Yang: concept, design, manuscript preparation, manuscript editing, and manuscript review. Xiaqing Wu: concept, design, clinical studies, manuscript editing, and manuscript review. Hua Li: literature search, clinical studies, manuscript editing, and manuscript review. Runji Zhou: experimental studies, manuscript editing, and manuscript review. Kai Guo: data acquisition, data analysis, manuscript editing, and manuscript review. Chunping Shang: literature search, data acquisition, manuscript editing, and manuscript review. Songhua Zhao: concept, design, data analysis, manuscript editing, and manuscript review. Mingyi Ma: concept, design, definition of intellectual content, manuscript editing, and manuscript review.
Acknowledgments
We are grateful to all study participants for their active engagement in this study. We wish to express our appreciation to the hand surgery clinician at the Affiliated Hospital of Southwest Medical University for his professional guidance throughout this study.
Ethics Statement
The research protocol was conducted in accordance with the Declaration of Helsinki guidelines, and approved by the Ethics Committee of the Affiliated Hospital of Southwest Medical University (Approval No. KY2023410).
Consent
Informed consent was obtained from all participating subjects.
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
The data that supports the findings of this study are available in the Supporting Information of this article.




