Nablus mask-like facial syndrome: Report of an atypical case with 8q21.3–q22.1 deletion
Abstract
Nablus mask-like facial syndrome (NMLFS) is a rare condition characterized by unique facial features, initially described in a 4-year-old boy from Nablus, Palestine. These features include expressionless facial appearance, tight facial skin, blepharophimosis, sparse eyebrows, and a flat nose. Genetic studies have identified a deletion of 8q22.1 as the cause of the syndrome, however while 26 patients have been reported with the deletion, only 13 displayed the characteristic facial features. Here we report on a 35-year-old male with 8q21.3–q22.1 deletion identified by whole exome sequencing and Chromosomal microarray analysis (CMA) that presents with typical and atypical features, including neurodevelopmental disorder, mild facial features, and myopathy, which has not been described in a patient with NMLFS to date. Further research will be required to understand the underlying pathogenetic mechanism of this rare genetic disorder.
1 INTRODUCTION
Nablus mask-like facial syndrome (NMLFS, OMIM #608156) is a very rare condition first described in a 4-year-old boy in the city of Nablus, Palestine by Teebi (2000). The hallmark feature of the syndrome is its characteristic expressionless facial appearance, with tight glistening facial skin, blepharophimosis, sparse arched eyebrows, upswept frontal hairline, and flat and broad nose, resembling a mask (Jain et al., 2010). In 2006, Shieh and colleagues determined that the syndrome is caused by a ~4 Mb deletion of 8q21.3–q22.1 (Shieh et al., 2006). To this day 26 patients with the deletion have been reported in the literature, but only 13 showed the characteristic facial features of the syndrome (Allanson et al., 2018). Here we report on an additional patient with 8q21.3–q22.1 deletion presenting typical and atypical features.
2 CASE REPORT
The patient is a 35-year-old male born at term from non-consanguineous parents after an uncomplicated pregnancy, with a birth weight of 3250 g, length of 50 cm, and head circumference of 34 cm. The perinatal period is reported to have been uncomplicated. He is the first of two children born to his parents. He presented with hypotonia, cryptorchidism, and laryngomalacia (Figure 1a). He sat at 6 months, walked at 2.5 years, and spoke his first words at 3.5 years. Conventional G-banded chromosome analysis revealed a normal male karyotype, 46, XY.

At the age of 9, he was diagnosed with attention deficit hyperactivity disorder (ADHD), learning disabilities, and atypical behavior (Figure 1b). Specifically, he banged his head, destroyed objects presented to him, drooled, and placed his hands in his mouth. Expert assessment documented moderate intellectual impairment. He had difficulty writing, and he did not play with children his age. EEG was normal and developmental assessment placed him in the lower normal range. He was treated with Ritalin (10 mg daily), which improved his hyperactivity disorder. A CT scan was performed, which did not reveal any pathogenic findings.
At age 18 he was referred to the Laboratory of Medical Genetics of the National and Kapodistrian University of Athens for clinical genetic evaluation, which noted moderate to severe psychomotor delay, laryngomalacia, myopia (6dpt) and keratoconus. Electromyography and brain Magnetic Resonance Imaging (MRI) were normal. At that time, his height was 162 cm, weight 57 kg, and head circumference 54 cm (3rd centile), denoting microcephaly. Additionally, he presented with hypertelorism, prominent nasal root, short philtrum, arched palate, teeth abnormalities, low set ears, prominent upper jaw, frontal upswept hairline, joint hyperextensibility, cryptorchidism, (for which he underwent surgery twice) and enlarged penis. He had asymmetrical fingers, with wide thumbs, and mild abnormalities of the fingerprints as well as mild interdigital webbing (Figure 1e,f), while his skin was described as “silky.” Also, he complained of constipation. Chromosome analysis was repeated and reconfirmed normal male karyotype and Fragile X molecular analysis was negative.
Electromyogram (EMG), at age 19, indicated mild diffuse loss of muscle fibers and subsequent muscle biopsy (at age 21) disclosed mild histopathological lesions consistent with myopathy presenting as contractures of the feet and ankle joints. Moderate insufficiency of the tricuspid valve was also indicated on echocardiogram.
A decade later, at age 33 (Figure 1c, d), the progression of his myopathy led to difficulties in walking and lifting objects. Due to deterioration of his neurological status, whole exome sequencing was performed. Although no pathogenic Single Nucleotide Variants (SNVs) were detected to explain the clinical phenotype, a deletion of the 8q21.3–q22.2 region was revealed requiring confirmation with an orthogonal method. Chromosomal microarray analysis was performed to confirm the finding and allow more accurate definition of breakpoints.
3 METHODS
3.1 DNA extraction
DNA was extracted from peripheral blood lymphocytes in EDTA using the QiaSymphony SP robotic system and the QIAsymphony DSP DNA Mini Kit. Quality and concentration of extracted DNA was determined with the NanoDrop 1000 UV–Vis spectrophotometer (Thermo Scientific) and the Qubit 3.0 fluorophotometer (Thermo Scientific).
3.2 Whole exome sequencing
Whole exome sequencing was performed on an Illumina NextSeq-500 sequencer with the capture-based IDT xGen Exome Research v2 library preparation kit (Integrated DNA Technologies, USA), covering the coding regions of ~19.000 genes (~34 Mbp). A total of 9.118.152.750 bases were sequenced from a total of 60.787.685 reads (99.71% mapped reads). A total of 30,785 exonic variants were identified. 99.3% of all coding regions had at least 25× coverage, while the mean coverage was at 111×. Bioinformatic analysis including alignment against GRCh37/hg19 version of the human genome and variant calling was performed with the VarSome Clinical platform (Saphetor, Switzerland). Variant annotation was performed with the use of available databases, scientific literature, and in silico variant effect prediction tools including 1000 Genomes, gnomAD (ExAC), UMD-predictor, Shift, PolyPhen, Mutation Taster, ClinVar, VarSome, OMIM, and HPO. Copy Number Variants from exome data were inferred using ExomeDepth (Plagnol et al., 2012), which provides an indirect estimate of Copy Number utilizing the coverage of the exonic regions (Tilemis et al., 2023).
3.3 Chromosomal microarray analysis
Chromosomal microarray analysis was performed using the high-resolution 4x180K G3 comparative genomic hybridization (CGH) + single nucleotide polymorphism (SNP) microarray platform (G5890A, Design ID 029830, Agilent Technologies, Santa Clara, CA, USA). The specific platform features a total of 110,712 oligonucleotide CGH probes annotated against National Center for Biotechnology Information (NCBI) Build 37/hg19 (UCSC GRCh37, Dec. 2013), covering the whole genome with a median CGH probe spacing of 25.3 kb as well as 59,647 SNP probes for the detection of copy-neutral loss of heterozygosity. The laboratory protocol was carried out according to the manufacturer's instructions (Agilent Oligonucleotide Array-Based CGH for Genomic DNA Analysis) and in short consisted of enzymatic digestion of the genomic DNA sample as well as a sex-matched reference DNA (Agilent Technologies), followed by differential labeling with Cy5 and Cy3 fluorescent dyes for sample and reference, respectively. The combined labeled DNA samples were applied to the microarray, left to hybridize for 24 h at 67 C, washed and scanned at 3 μm resolution on the SureScan Dx microarray scanner (Agilent Technologies). The tiff images were extracted and analyzed using the Agilent Feature Extraction software and the CytoGenomics v.5.0.1 software suite (Mitrakos et al., 2020). The ADM-1 aberration detection algorithm was utilized with the minimum number of probes required for a call set to four and the log2ratio threshold for duplications and deletions set to ±0.25. For the evaluation and classification of Copy Number Variants (CNVs) the literature as well as available databases were used, including Franklin, Database of Genomic Variants (DGV), DECIPHER, and ClinVar. All variants were classified according to American College of Medical Genetics and Genomics (ACMG) and ClinGen guidelines (Richards et al., 2015; Riggs et al., 2020). Informed consent was obtained from the patient's legal guardians for publication of this article.
4 RESULTS
Whole exome sequencing (WES) identified a heterozygous deletion of ~8.8 Mb of 8q21.3–q22.2 (89179899-97978274, GRCh37), which encompassed 60 genes and was classified as pathogenic according to the ACMG and ClinGen criteria 1A, 2A, 2H, 3C, and 4L (Figure 2). CMA confirmed the presence of the heterozygous deletion at 8q21.3—q22.1 region and further defined its size (8.87 Mb) and breakpoints (89148404-98017865, GRCh37, Figure 3, Table 1). The newly defined deletion was also classified as pathogenic based on the ACMG and ClinGen criteria 1A, 2A, 2H, 3C, and 4L. No other causative pathogenic or likely pathogenic SNVs or CNVs were detected either in the homologous alleles of genes included in the deletion or other loci as investigated by WES and CMA.
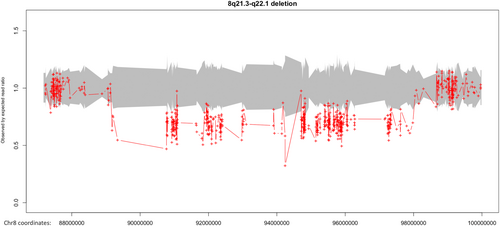
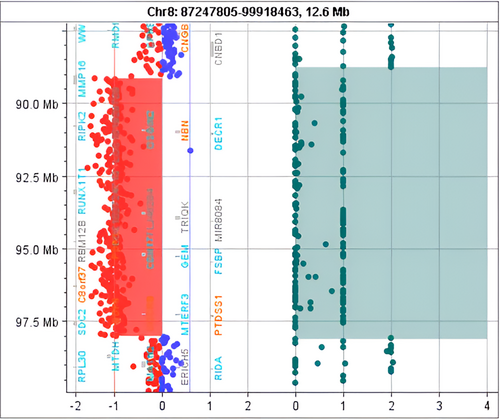
| ISCN description/size | Genes |
|---|---|
arr[GRCh37]8q21.3q22.1(89148404_98017865) × 1 8.87 Mb |
MMP16, LOC101929709, RIPK2, OSGIN2, NBN, DECR1, CALB1, LINC00534, LINC01030, TMEM64, NECAB1, C8orf88, PIP4P2, OTUD6B-AS1, OTUD6B, LRRC69, MIR4661, SLC26A7, RUNX1T1, LOC102724710, FLJ46284, TRIQK, MIR8084, C8orf87, LINC00535, FAM92A, RBM12B, RBM12B-AS1, TMEM67, MIR378D2, PDP1, CDH17, GEM, RAD54B, FSBP, VIRMA, LOC100288748, ESRP1, DPY19L4, INTS8, CCNE2, NDUFAF6, TP53INP1, MIR3150BHG, MIR3150B, MIR3150A, PLEKHF2, LINC01298, C8orf37, C8orf37-AS1, LOC100500773, GDF6, UQCRB, MTERF3, PTDSS1, LOC102724804, SDC2, CPQ, LOC101927066 |
5 DISCUSSION
NMLFS, also known as blepharo-naso-facial syndrome, is a rare genetic disorder characterized by distinct facial characteristics. The hallmark features of the syndrome have been well established and include blepharophimosis, tight-appearing glistening facial skin, an abnormal hair pattern with an upswept frontal hairline, sparse arched eyebrows, flat and broad (“bulbous”) nose, long philtrum, distinctive ears, and a happy demeanor (Jain et al., 2010). However, the clinical presentation of NMLFS varies widely among patients, with additional features in most of the patients reported including micrognathia, microcephaly, intellectual disability and/or developmental delay, and cryptorchidism, while isolated features may include genital anomalies, joint anomalies, submucous cleft palate, cleft lip, abnormal teeth, inguinal hernias, interdigital webbing, and camptodactyly (Allanson et al., 2018).
In 2000 Teebi was the first to describe a boy with a “strikingly unusual facial appearance,” including a long face, upswept frontal hairline, tight glistening facial skin, blepharophimosis, hypertelorism, and sparse eyebrows, among other features. Subsequent reports identified similarities between NMLFS and blepharo-naso-facial syndrome, with features such as telecanthus, nasal abnormalities, and intellectual disability. However, the genetic etiology remained unclear until Shieh and colleagues in 2006 identified a microdeletion in the 8q21.3–q22.1 region as the cause of NMLFS, a finding further supported by later studies on additional patients with NMLFS.
The clinical spectrum expanded with the report of cases exhibiting additional features such as microcephaly, craniosynostosis, joint contractures, and genital abnormalities. However, not all patients with 8q21.3q22.1 deletions presented typical NMLFS features, suggesting a high degree of variable expressivity. Of note, in 2010 Jain and colleagues (Jain et al., 2010) reported a patient with speech delay and autism spectrum disorder but lacking typical NMLFS features despite having an 8q22.1 deletion. This finding highlighted the complexity of the syndrome and suggested the involvement of other genetic factors. The observation of further patients with the deletion and neurodevelopmental disabilities but not the characteristic facial appearance of the syndrome (Debost-Legrand et al., 2013; Jamuar et al., 2015; Overhoff et al., 2014) led to the hypothesis that a deletion of 8q22.1 is necessary but not sufficient to produce NMLFS phenotype (Allanson et al., 2012).
These observations suggested that the underlying pathogenetic mechanism might be more complex than simple haploinsufficiency of a gene or genes in the deleted region. Allanson and colleagues proposed possible models for the molecular pathogenesis of NMLFS. While a specific gene or genes in the 8q22.1 region is probably implicated, its expression may depend on an interaction between the 8q22.1 microdeletion and other genetic variants within the critical region (one-locus model) as well as common or rare variants outside the critical region (two-locus model). Epigenetic effects and gene–environment interactions were also suggested as potential contributors to the clinical heterogeneity observed (Allanson et al., 2018).
Here we report on an additional adult patient with (NMLFS, #608156) with an 8.87 Mb deletion of 8q21.3q22.1 who presents both typical and atypical phenotypic features. Apart from the 8q21.3q22.1 deletion, no other pathogenic or likely pathogenic SNVs or CNVs associated with the patient's phenotype were identified. The deletion was not detected through conventional G-banding analysis, highlighting the higher diagnostic potential offered by newer methods, including chromosomal microarray analysis, and whole exome sequencing, toward the detection of causative variants.
Our patient shows most of the hallmark features of the syndrome as he presents with hypotonia, cryptorchidism, laryngomalacia, ADHD, developmental delay, learning difficulties, behavioral disorder, hypertelorism, nasal root protrusion, short philtrum, arched palate, teeth abnormalities, low set ears, upper jaw protrusion, cryptorchidism and enlarged penis, elasticity of the joints and fingers and upswept hairline, mild abnormalities of the fingerprints, and mild interdigital webbing. His skin was described as “silky”; however, he lacks severe facial features of the syndrome while he also presents with symptoms and signs of myopathy with abnormal EMG findings. Although joint contractures were previously reported in patients with NMLFS, myopathy is not yet considered an established feature (Raas-Rothschild et al., 2009; Shieh et al., 2006), however it is possible that myopathy is a previously unappreciated or underappreciated manifestation of the syndrome, with the hallmark features (expressionless face, ptosis, blepharophimosis, joint contractures and smooth tight skin) observed in most cases of Nablus syndrome being signs of prenatal onset myopathy, that would have been identified by EMG had such studies been carried out. To the best of our knowledge no report of a patient with Nablus syndrome and abnormal EMG findings exists in the literature. A later onset of myopathic symptoms in patients with the 8q22.1 deletion may also be postulated, although the few reports concerning adults with the deletion (mainly the parents of affected offsprings) do not provide any such evidence (Allanson, 2002).
The minimal region of overlap in all patients reported to date has been narrowed down to a 1.84 Mb region (Figure 4) at 8q22.1 (94.43–96.27 Mb, hg19), encompassing more than 20 genes, including coding genes, noncoding RNAs and microRNAs (Allanson et al., 2018; Overhoff et al., 2014). The main coding genes included in the region comprise FAM92A1, RBM12B, TMEM67, PDP1, CDH17, GEM, RAD54B, FSBP, KIAA1429, ESRP1, DPY19L4, INTS8, CCNE2, NDUFAF6, TP53INP1, PLEKHF2, and C8orf37, of which 6 are associated with pathogenic phenotypes in OMIM (Allanson et al., 2018). The region also includes LINC00535, cORF69, LOC100288748, MIR378D2, MIR3150B, and MIR3150A.
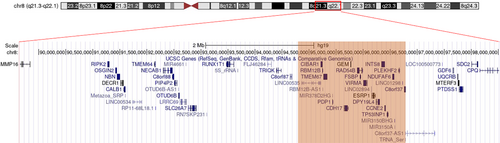
Most patients reported thus far carry a deletion of ~3.3–4.2 Mb, while a larger 7.27 Mb deletion (89.07–96.34, hg19) was observed in a case described by Raas-Rothschild and colleagues (Raas-Rothschild et al., 2009) with no symptoms associated with myopathy. An assumption that genes related to the myopathy phenotype concern GDF6, UQCRB, MTERF3, PTDSS1, SDC2, and CPQ which lie in the remaining nonoverlapping region could be posed, however none of these genes has been associated with a myopathy phenotype to date. Additional genetic variants such as variants in noncoding regions failing detection by WES and CMA cannot not be excluded.
6 CONCLUSION
NMLFS is a rare genetic disorder characterized by a constellation of distinct facial features resulting from microdeletions in the 8q21.3q22.1 region. The clinical presentation varies widely among individuals, with some showing typical features while others exhibit additional abnormalities or lack characteristic facial features altogether. Our patient has dysmorphic features, neurodevelopmental disorder, and myopathy. Myopathy has not yet been reported as a consistent feature or directly associated with NMLFS. However, given the rarity of the syndrome, there may be cases where additional symptoms or comorbidities, including myopathy, could be present but unappreciated. A more thorough examination of patients with 8q22.1 deletions, including EMG studies, might be necessary to identify possible myopathic features. The study of larger patient cohorts with comprehensive approaches will be needed to reveal the pathogenetic mechanisms underlying the variable expressivity and incomplete penetrance observed for the syndrome, shedding light on the manifestation of complex genetic disorders.
AUTHOR CONTRIBUTIONS
Anastasios Mitrakos: Conceptualization; formal analysis; data curation; methodology; writing—original draft; writing—review and editing. Kyriaki Kekou: Formal analysis; data curation; methodology; writing—review and editing. Faidon-Nikolaos Tilemis: Formal analysis; software; data curation; methodology; and writing—original draft. Maria Svingou: Formal analysis; data curation; and methodology. George Papadimas: Formal analysis and clinical evaluation. Christalena Sofocleous: Formal analysis; writing—review and editing. Joanne Traeger-Synodinos: Project administration; writing—review and editing. Maria Tzetis: Supervision; project administration; writing—review and editing.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




