Molecular autopsy in Chinese sudden cardiac death in the young
Abstract
Inherited cardiovascular conditions are significant causes of sudden cardiac death in the young (SCDY), making their investigation using molecular autopsy and prevention a public health priority. However, the molecular autopsy data in Chinese population is lacking. The 5-year result (2017–2021) of molecular autopsy services provided for victims of SCDY (age 1–40 years) was reviewed. The outcome of family cascade genetic screening and clinical evaluation was reviewed. A literature review of case series reporting results of molecular autopsy on SCDY in 2016–2023 was conducted. Among the 41 decedents, 11 were found to carry 13 sudden cardiac death (SCD)-causative genetic variants. Likely pathogenic (LP) variants were identified in the DSP, TPM1, TTN, and SCN5A genes. Cascade genetic testing identified four family members with LP variants. One family member with familial TPM1 variant was found to have hypertrophic cardiomyopathy upon clinical evaluation. This study provided insight into the genetic profile of molecular autopsy in a Chinese cohort of SCDY. The detection of important SCD-causative variants through molecular autopsy has facilitated family cascade screening by targeted genetic testing and clinical evaluation of at-risk family members. A literature review of the current landscape of molecular autopsy in the investigation of SCDY was conducted.
1 INTRODUCTION
Sudden cardiac death in the young (SCDY) has a devastating impact on families and societies, making it a matter of public health concern. The reported incidence of sudden cardiac death (SCD) in children and young adults ranges from 0.91–8.7 per 100,000 person-years (Ha et al., 2020; Lynge et al., 2019; Wong et al., 2019). Inherited cardiovascular condition accounts for a majority of SCDY (Lynge et al., 2019). Among the decedents, 40% of causes of death are unascertained at postmortem examination and were presumed to be due to sudden arrhythmia death syndrome (SADS). In addition, identifiable inheritable structural causes such as hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy, and arrhythmogenic cardiomyopathy (ACM), attributed to around 16% of cases (Bagnall et al., 2016). With molecular autopsy, clinically relevant cardiac genetic variants were identified in 27% of SCDY (Bagnall et al., 2016). Through family cascade genetic screening, with a yield of up to 33%, early assessment and intervention can be provided to at-risk family members to facilitate prevention of SCDY. (Van Der Werf et al., 2010). Investigation of SCDY should be made a public health priority, due to the combined prevalence of inherited cardiac diseases of at least 1:500, the years of potential life loss, and the significant impact on the family and community (Stiles et al., 2021).
Molecular autopsy services in Hong Kong are currently provided by the Clinical Genetics Service Unit (CGSU) at Hong Kong Children's Hospital, with referrals received from the Forensic Pathology Service (FPS) of the Department of Health. In cases where inheritable cardiovascular conditions are suspected, family members are offered cascade genetic screening and referred to cardiac units for further evaluation. With a lack of molecular autopsy data in Chinese population, this report serves to describe the genetic landscape of molecular autopsy in a cohort of Chinese patients with SCDY.
2 MATERIALS AND METHODS
SCD refers to death that occurs within 1 h of onset of symptoms in witnessed cases, and within 24 h of last being seen alive when it is unwitnessed. Sudden unexplained death syndrome is defined as unexplained sudden death occurring in an individual older than 1 year. SADS is defined as unexplained sudden death occurring in an individual older than 1 year with negative pathological and toxicological assessment, and it is synonymous with autopsy-negative sudden unexplained death (Stiles et al., 2021). SCDY refers to SCD victims who died at the age of 1–40 years.
For the SCDY decedents, molecular autopsy request was made to CGSU when gross autopsy revealed either cardiomyopathy or no anatomical cause of death, and with a negative toxicology screening. Molecular autopsy would not be performed if the gross autopsy showed coronary artery diseases, anomalous coronary arteries, aortic dissection, congenital heart disease, and myocarditis, in which the genetic components were negligible (Stiles et al., 2021). A search was conducted in the CGSU database, and a retrospective cohort review was performed for cases with gross autopsy finding of cardiomyopathy or molecular autopsies conducted in CGSU for decedents of SCDY at age 1–40 years, from August 2017 to September 2021. Victims with cardiomyopathy secondary to chemotherapy, alcoholism, myocarditis, environmental toxin exposure, or nutrient deficiency were excluded. Only Chinese victims were included for analysis.
2.1 DNA preparation method
DNA was extracted from peripheral blood or tissue sample taken during autopsy. Genomic DNA was extracted using Maxwell RSC Whole Blood DNA Kit from Promega, following the manufacturer's protocol. DNA concentration was measured using a spectrophotometer (NanoDrop 2000).
2.2 Library preparation and NGS
Next generation sequencing (NGS) employing the TruSight Cardio gene panel kit was used before 2019, and exome sequencing was adopted after 2019 in CGSU for molecular autopsy analysis.
The TruSight Cardio gene panel kit, provided by Illumina in San Diego, CA, included 174 genes associated with hereditary cardiac diseases that were most affected by a genetic predisposition. Genes included in the panel are shown in Table S1 (http://www.illumina.com/cardio). The coding and splice junction regions of the target genes were enriched by using target capture system by Illumina. The targeted regions were then sequenced simultaneously through massively parallel sequencing on Illumina platform with paired-end reads.
For exome sequencing library preparation, medical exome was employed, and analysis was made with filtering according to genomic England panel app SCD panel (up to 261 genes to date). The medical exome was enriched by using capture system by Agilent Sureselect V6. The targeted regions were sequenced simultaneously by massively parallel sequencing on NextSeq 500 with paired-end reads.
2.3 Bioinformatic analysis
Bidirectional sequence reads were assembled and aligned to the human reference genome based on human genome build GRCh37/USCS hg19 by ELSA aligner and variant caller, followed by bioinformatics analysis. The system used includes Helicube version 2.2.1, Database.bio version 2.2.7, and ELSA version 1.5.4a. Sanger sequencing was utilized to confirm all reported potentially pathogenic variant(s) with low-quality metrics identified in the bioinformatic pipeline. The variants were filtered and prioritized by considering (1) the pattern of inheritance within each family; (2) variant filtering to a minor allele frequency (MAF) consistent with the mode of inheritance and disease frequency, generally these are <=1% of autosomal recessive conditions or <=0.1% of autosomal dominant conditions in the Exome Aggregation Consortium (exac.broadinstitute.org); and (3) databases (including public version of HGMD, ClinVar, LSDBs, NHLBI Exome Sequencing Project, 1000 Genomes, and ExAC), published literature and computational tools (Alamut visual 2.10). Variants were classified according to the American College of Medical Genetics and Genomics (ACMG) standards and guidelines. Benign/likely benign variants were not reported (Richards et al., 2015).
2.4 Information on family cascade genetic screening and clinical evaluation
If pathogenic (P) or likely pathogenic (LP) variants were identified, family cascade genetic screening would be offered to the first-degree family members, using targeted Sanger sequencing. For decedents with variants of uncertain significance (VUS) identified, genetic testing for first-degree family members might be performed after genetic counseling for the purpose of variant reclassification. Autopsy reports were retrieved with permission from the coroner office for detailed review, and a chart review was conducted. For victims identified with positive gross autopsy, and/or found to have pathogenic variants or VUS, their next-of-kin were contacted as part of routine follow-up service. Additional clinical information about the family members, particularly regarding the progress and results of their subsequent cardiovascular evaluation were collected.
2.5 Literature review on SCDY
A systematic search in PubMed (2016–2023) was conducted using the keywords “molecular autopsy,” “postmortem genetic test,” and “sudden unexplained death.” Retrospective and prospective case series, published in English, with SCDY victims having undergone molecular autopsy were all included. Full articles were retrieved with detailed analysis of the followings: the study design, number of subjects included in each study, age range of subjects, and the methodology employed for the molecular autopsy. If a gene panel was used, the number of genes included in the panel analysis was recorded. Specifically, the MAF cutoff used in the identification of variants was searched and recorded if available. The adherence to the ACMG guidelines for variant determination was carefully assessed in each publication. The findings and yield of molecular autopsy, categorized as P/LP and VUS, were summarized. The top five variants identified in each publication were quoted, highlighting the most frequently observed variants.
2.6 Statistical analysis
The genetic yield of molecular autopsy was calculated based on the number of decedents identified to have genomic causes. If both P/LP variants and VUS were found in a decedent, the P/LP variants were regarded to be the molecular explanation. Descriptive statistics were analyzed using SPSS Statistics, Version 24.0 (Armonk, NY: IBM Corp.) and Microsoft Excel software. Categorical variables were expressed as count and percentage, whereas continuous variables were expressed as median and interquartile ranges (IQR). A p-value of <0.05 was considered statistically significant.
2.7 Editorial policies and ethical considerations
The study was approved by the Institutional Review Board of the Hong Kong Children's Hospital (PAED-2022-028) and the Department of Health (LM 345/2022), Hong Kong, China. The rights and privacy of the participants were protected, and the research was conducted in an ethical and responsible manner, adhering to the relevant guidelines and regulations governing human research. Informed consent was obtained from surviving participants.
3 RESULTS
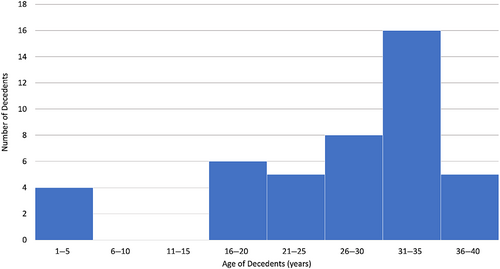
A total of 44 decedents aged 1–40 years were identified. The age distribution is shown in Figure 1. Most sudden death victims were young adults from 25 to 40 years old. There were a few pediatric cases, with age from 1 to 5 years, but none was found in the age group from 6 to 15 years. The median age was 31.4 years (IQR 26.2–36.1 years). All cases were referred from FPS, except two of which were referred from anatomical pathology units of public hospitals. Thirty-eight of them were male (86.4%). One Indonesian and one Pakistani were excluded. One 32-year-old victim was found to have HCM on autopsy examination, but extracted DNA quality was suboptimal to undergo molecular autopsy. Therefore, molecular autopsy was performed on a total of 41 decedents. Autopsy reports were reviewed, and none of these cases had coronary artery diseases, anomalous coronary arteries, aortic dissection, congenital heart disease, and myocarditis as the leading causes of death.
3.1 Molecular findings with autopsy findings of cardiomyopathy
Eleven decedents with autopsy findings of cardiomyopathy were referred. Table 1 shows the gross autopsy results with their corresponding molecular findings. Three LP variants and three VUS were identified in 5/11 decedents (45.5%) who had undergone molecular autopsy. These six variants involved five different SCD-related genes. The yield of LP variants was 27.3% (3/11 decedents) and that of VUS was 18.2% (2/11 decedents).
| Sudden cardiac death victims with positive autopsy findings of cardiomyopathy | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Decedent | Age | Sex | Activity at time of death | Gross autopsy | Gene | Nucleotide change | Protein change | ACMG classification | Coding effect | Documented VT/VF |
| 1 | 16 | M | Rest | HCM | Negative | |||||
| 2 | 18 | M | Sleep | DCM | Negative | |||||
| 3 | 25 | M | Unknown | ACM | Negative | |||||
| 4 | 29 | M | Rest | HCM | CACNA1C | c.5242G>A | p.Gly1748Ser | VUS | Missense | |
| 5* | 30 | M | Light activity | ACM | DSP | c.4003C>T | p.Gln1335Ter | LP | Nonsense | Yes |
| 6 | 31 | M | Running | HCM | TPM1 | c.644C>T | p.Ser215Leu | LP | Missense | Yes |
| 7 | 32 | M | Postexercise | HCM | TTN | c.10840G>T | p.Glu3614Ter | LP | Nonsense | |
| ACTN2 | c.2425G>A | p.Val809lle | VUS | Missense | ||||||
| 8 | 32 | M | Rest | ACM | Negative | Yes | ||||
| 9 | 34 | M | Unknown | ACM | Negative | |||||
| 10 | 35 | M | Sleeping | DCM | Negative | |||||
| 11 | 35 | F | Rest | LVNC | DSP | c.449G>A | p.Arg150Gln | VUS | Missense | |
| Sudden cardiac death victims with negative autopsies, in which variants in SCD genes were identified | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Decedent | Age | Sex | Activity at time of death | Gene | Nucleotide change | Protein change | ACMG classification | Coding effect | Documented VT/VF |
| 12 | 17 | M | Light activity | MIB1 | c.2370_2371insCT | p.Lys791Leufs*19 | VUS | Nonsense | |
| 13 | 18 | M | Postexercise | AKAP9 | c.5779T>C | p.Tyr1927His | VUS | Missense | |
| 14* | 29 | M | Unknown | TNNT2 | c.100C>A | p.Gln34Lys | VUS | Missense | |
| 15 | 31 | M | Sleep | SCN5A | c.4282G>T | p.Ala1428Ser | LP | Missense | Yes |
| 16* | 32 | M | Sleep | ANK2 | c.1627G>A | p.Val543Met | VUS | Missense | |
| 17* | 35 | M | Unknown | RYR2 | c.1162C>A | pGln388Lys | VUS | Missense | |
| AKAP9 | c.2696A>C | p.Glu899Ala | VUS | Missense | |||||
- Abbreviations: ACM, arrhythmogenic cardiomyopathy; ACMG, American College of Medical Genetics and Genomics; DCM, dilated cardiomyopathy; HCM, hypertrophic cardiomyopathy; LP, likely pathogenic variant; LVNC, left ventricular noncompaction cardiomyopathy; SCD, sudden cardiac death; VF, ventricular fibrillation; VT, ventricular tachycardia; VUS, variant of uncertain significance.
- * Exome-based testing.
3.2 Molecular findings with negative gross autopsy
Among the 30 decedents with negative gross autopsies, one LP variants and six VUS were identified in six decedents (Table 1), making a yield of 3.3% for LP variants and 16.7% for VUS. The only LP variant found was a SCN5A variant.
3.3 Sudden death related to ventricular arrhythmia
Eight decedents had documented ventricular arrhythmia or received defibrillations before their death (decedents 5, 6, 9, 15, Table 1). Among these, two victims had ACM and one had HCM, with DSP and TPM1 variants identified, respectively. One victim was identified to carry a SCN5A LP variant. Two decedents had a family history of SCD.
3.4 Family cascade genetic screening and clinical evaluation
The pathogenicity determination of LP variants identified in our cohort was described in Section S1. For decedents with LP genetic variants identified, the outcome of family cascade genetic screening and clinical evaluation of at-risk family members was illustrated in Figure S1. Family cascade genetic screening was performed on 16 individuals, except for decedent 5 whose next-of-kin failed to be contacted after availability of molecular autopsy results. Four individuals were identified to carry the LP variants (one TPM1 and three SCN5A).
Six family members were found to carry VUS on targeted genetic screening (two ACTN2, MIB1, AKAP9, ANK2, and RYR2/AKAP9), with an aim for phenotype–genotype correlations for the detected variants. However, none of these family members had abnormal findings upon clinical evaluation.
Overall, 12 first-degree relatives (out of 20) received clinical evaluation. All cardiovascular investigations, including electrocardiogram and echocardiography, yielded negative results, except for the father of a victim with TPM1 LP variant. He was found to have findings consistent with HCM on echocardiography.
4 DISCUSSION
This study summarized the genetic landscape of SCDY for the Chinese population, as a review of the largest cohort of Chinese SCDY victims who had undergone molecular autopsy.
Overall, the percentage of decedents identified with P/LP variants and VUS was 9.8% and 17.1%, respectively. The yield of LP variants was higher in cases with a postmortem diagnosis of established cardiomyopathy compared with cases without structural causes identified, which has been proven in a previous study (Lahrouchi et al., 2019). PubMed database was searched to identify all case series in the use of molecular autopsy in SCDY after 2016, and Table 2 summarized the results (Anastasakis et al., 2017; Bagnall et al., 2016; Cann et al., 2016; Christiansen et al., 2016; Fadoni et al., 2022; Hata et al., 2016; Hellenthal et al., 2016; Iglesias et al., 2021; Lahrouchi et al., 2017; Larsen et al., 2019; Li et al., 2023; Mak et al., 2019; Methner et al., 2016; Neubauer et al., 2018; Nunn et al., 2015; Ripoll-Vera et al., 2021; Rohrer et al., 2023; Sanchez et al., 2016; Scheiper et al., 2018; Scheiper-Welling et al., 2022; Shanks et al., 2018; Shanks et al., 2017; Thomas et al., 2018; Torkamani et al., 2016; Votýpka et al., 2023; Wang et al., 2023; Webster et al., 2021; Williams et al., 2019).
| Year of publication, author | Study design | Number of subjects | Country | Age (year) | Top variants identified | Methodology, MAF cutoff | ACMG rule used | Findings and yield | Remarks |
|---|---|---|---|---|---|---|---|---|---|
| 2016, Bagnall et al. | Pros | 490 SCD, 113 SUDY (M:F = 18:7) | Australia/NZ | 1–35 | RYR2, PKP2, ANK2, SCN5A, MYBP3 | Gene panel (59 genes)/WES/CNV analysis, MAF <0.1% | No | P: 27%, Familial diagnosis: 13% | Yield dropped to 7.1% on revisit |
| 2016, Sanchez et al. | Pros | 119 SUD, 32 CM (M:F = 77.2:22.8) | Spain | 0–50 | TNNI3, SCN5A, MYBP3, PKP2, TTN, CSRP3 | Gene panel/Sanger sequencing, MAF <0.1% | No | SUD: P/LP: 41.2%, VUS: 50.76%, CM: P/LP: 33.87%, VUS: 62.9% | |
| 2016, Methner et al. | Retro | SUD, 346 (0–1 y) (M:F = 60.1:39.9), 83 (1–40 y) (M:F = 62.7:37.3) | US | 0–40 | DSP, SCN3B, MYL2, SCN5A, SCN1B, MYBP3 | Gene panel (64 genes) | No | P: 7% | |
| 2016, Christiansen et al. | Pros | 61 (M:F = 7:3) | Denmark | 1–50 | RYR2, KCNH2, SCN5A, MYBP3, KCNQ1 | Gene panel (100 genes), MAF <1% | No | Variants with functional effect: 34% | |
| 2016, Nunn et al. | Retro | 59 (M:F = 19:6) | UK, Spain, Denmark | 1–55 | SCN5A, RYR2, DSC2, TTN, MYOT, GJA5 | Targeted exome analysis (135 genes), MAF <0.02% | No | Very rare P/LP variants (MAF <=0.02%): 14%, rare P/LP variants (MAF <=0.5%): 29% | |
| 2016, Torkamani et al. | Retro | 25 (M:F = 4:1) | US | 0–45 | HCN4, MYH6, PKP2, TRPM4 | WES | No | P: 40% | |
| 2016, Hata et al. | Pros | 25 (M:F = 21:4) | Japan | 18–50 | KCNQ1, KCNH2, MYH7, MYBP3, DSG2 | Gene panel (70 genes), MAF <0.5% | No | 56% | |
| 2017, Lahrouchi et al. | Pros | 302 (M:F = 13:9) | UK, Denmark, NZ, Netherlands | 1–64 (78%: <35) | KCNH2, RYR2, KCNQ1, SCN5A, TTN | Gene panel (77 genes), MAF <0.001% | Yes | LP/P: 13%, VUS: 42% | Combined yield of clinical screening |
| 2017, Cann et al. | Retro | 46 SUD, 50 CM (M:F = 13:10) | Scotland | 1–40, CM: 2–67 | KCNQ1, KCNH2, SCN5A, RYR2 | Gene panel (18 genes—disease specific) | No | SUD: P/LP 15%, CM: P/LP 18% | |
| 2017, Hellenthal et al. | Pros | 10 (M:F = 1:1) | Germany | 19–40 | KCNH2, CACNA1C, TNNT2 | Gene panel (174 genes) | Yes | P: 30%, VUS: 50% | |
| 2018, Neubauer et al. | Pros | 34 (M:F = 13:4) | Switzerland | 1–63 (82%: 1–40) | RYR2, ACAD9, AKAP9, KCNE5, SEMA3A | WES (192 genes), MAF <=1% | No | P: 29.4% | |
| 2018, Shanks et al. | Pros | 28 (M:F = 16:9) | US | 1.3–38 | DES, MYH7, RYR2, PLN, BAG3 | WES (Trio) (99 gene), MAF <0.005% | Yes | P/LP: 21.3%, VUS: 21.3%, autopsy positive: 31% | |
| 2018, Shanks et al. | Pros | 25 (M:F = 17:8) | Illinois, US | 1–40 | SCN5A, RYR2, NEBL, MYH7, TTN, MYH6, DES, DSP | WES, MAF <=0.005% (99 genes) | Yes | P/LP: 28%, VUS: 36%, clinically actionable variants: 14% | Ultrarare variant |
| 2018, Anastasakis et al. | Pros | 20 | Greece | 1–35 | PKP2, KCNH2, RYR2, TPM1 | Gene panel | No | Yield of clinical screening—all found causative mutation: 25% | Clinical guided genetic screening |
| 2018, Scheiper et al. | Retro | 9 (3 autopsy positive) (M:F = 5:4) | Germany | 1–45 | TTN | Gene panel (96 genes), MAF <0.2% | Yes | VUS: 66.7% | |
| 2019, Mak et al. | Pros | 21 (M:F = 18:3) | HK, China | 5–40 | DSC2, AKAP9, RYR2, CACNA1C, SCN5A, JUP | Gene panel (35 genes), MAF <0.1% | No | Clinical relevant P: 29% | |
| 2019, Larsen et al. | Retro | 70 (M:F = 7:3) | Denmark | 1–40 | CACNA1C, KCNH2, KCNQ1, RYR2, GAA, MYH7, TTN, SCN5A, PKP2, LMNA, MYBPC3, MYH6 | Gene panel (100 genes), MAF <0.05% | Yes | P/LP: 16%, VUS: 3% | |
| 2019, Williams et al. | Retro | 73: Arrhythmia/ CM | NY, USA | <35: 120 | MYBP3, TTN, KCNH2, RYR2, DSP | Gene panel (95 genes) | Yes | P/LP: 20.3%, VUS: 47.9% | |
| 2021, Iglesias et al. | Pros | 31 (M:F = 25:6) | Switzerland | 1–50 | MYBP3 | WES (194–380 genes), MAF <0.02% | Yes | P/LP: 6.46%, VUS: 61.29% | Selective autopsy cases |
| 2021, Ripoll-Vera et al. | Pros | 30 SUD, (M:F = 2:1) | Spain | <=50 | RYR2, SCN5A, KCNH2, MYBP3 | Gene panel (194–380 genes), MAF <0.02% |
Yes | P/LP: 26.6%, VUS: 43.3% | |
| 2021, Webster et al. | Pros | 103, 44 SUD (M:F = 3:1) | US | 1–44 | KCNH2, KCNQ1, MYL3, TNNI3, TTN | WGS (118 genes), MAP <0.2% | Yes | P/LP: 12.6% (SUD 11%; CM 17%), VUS/P/LP: 31.3% | |
| 2022, Scheiper-Welling et al. | Retro | 56 SCD (M:F = 19:9) | Germany | 1–50 | KCNH2, SCN5A, KCNQ1, KCNH2, MYBPC3, VCL, JUP | Gene panel (174 genes), MAF <0.2% | Yes | P/LP/VUS potentially pathogenic: 12% (51 VUS in 56 subjects) | |
| 2022, Fadoni et al. | Retro | 16 HCM (all male) | Portugal | 1–50 | MYBPC3, MYH7 | Panel test (40 genes), MAF <=5% | Yes | P/LP: 37.5% | |
| 2023, Rohrer et al. | Retro | 18 SUD (M:F = 13:5) | US | 2 months–49 | TTN, SCN5A, KCNH2, RYR2 | Gene panel (113 genes) and WES | Yes | P/LP: 38.9%, VUS: 38.9% | |
| 2023, Wang et al. | Retro | 27 SUD (M:F = 25:2) | China | 18–40 | TTN, SLC25A3 | Gene panel (255 genes), MAF <0.1% | Yes | P/LP: 51.9%, VUS: 100% | |
| 2023, Votýpka et al. | Retro | 100 SCD (M:F = 71:29) | Czech | 1–59 | TTN, FLNC, RYR2, COL3A1 | Gene panel (100 genes) | Yes | P/LP: 22.0% | |
| 2023, Li et al. | Retro | 16 CM (M:F = 12:4) | China | 22–70 | DSP, TTN, LMNA, SLC22A5, RYR2, ALMS1 | Gene panel (82 genes) | Yes | P/LP: 37.5% |
- Abbreviations: ACMG, American College of Medical Genetics and Genomics; CM, cardiomyopathy; MAF, minor allele frequency; NZ, New Zealand; P/LP, pathogenic/likely pathogenic variants; Pros, prospective; Retro, retrospective; SCD, sudden cardiac death; SUD, sudden unexplained death; SUDY, sudden unexplained death in the young; VUS, variants of uncertain significance.
There were 14 prospective and 13 retrospective cases series. For prospective studies, a total of 954 sudden death victims with potential inheritable cardiovascular conditions were investigated. The age and gender distribution of these reported cases series was comparable with our findings, with a higher number of sudden death victims in the young adult groups (25–40 years old) and male gender.
The yield of molecular autopsy varied across the published prospective case series (P/LP: 6.5%–56%; VUS: 21.4–62.9%). The most prevalent SCD genes identified were RYR2, KCNH2, SCN5A, MYBPC3, TTN, KCNQ1, and MYH3. The wide range of reported yield was attributed to heterogeneity in methodology of variant classification, cutoff of MAF chosen for variant filtering, and the selected gene panel coverage. When compared with the latest prospective US cohort by Webster et al., 2021, the yield of our cohort was comparable with their reported yield of 31.3%, though the yield for SADS was relatively lower in our cohort (3.1%).
The yield of VUS was nearly double that of LP variants in our cohort. They were not clinically actionable variants, and their causative role among the SCDY cases remained speculative. On review of the reported case series of molecular autopsies (Table 2), the variant pick-up rate was higher if more targeted genes were involved, but the proportion of VUS was also higher, up to a ratio of VUS:P/LP variants of 9.48 in the study by Iglesias et al., 2021. Therefore, it is recommended that SCDY genomic evaluation should include only genes where there are robust genotype–phenotype associations, so as to avoid VUS interpretation in “weak” genes (Stiles et al., 2021). Latest consensus statement on the state of genetic testing for cardiac diseases also advised against a hypothesis-free genetic testing using exome or genome sequencing in a decedent with SCDY (Wlide et al., 2022).
In clinical practice, VUS should not be used in the risk assessment of relatives of SCDY. Nevertheless, sometimes clinical evaluation of family members and family segregation analysis is imperative for the correct classification of SCD variants, especially in the absence of phenotype information of decedents, for example, negative autopsy findings. A combined genotype–phenotype approach improved genetic assessment of sudden death. Combining molecular autopsy with clinical evaluation in surviving family members has been proven to increase the diagnostic yield from 26% to 39% (Lahrouchi et al., 2017; Webster et al., 2021). The segregation information is not only part of criteria for variant classification but also helps to determine the clinical relevance of the identified variants to the family, especially if VUS are identified. (Fellmann et al., 2019). Having said that, testing family members for VUS has to be done with caution. Pre- and post-test genetic counseling are essential in order not to cause unnecessary anxiety. And, the genetic test should not be done without proper, and preferably prior, clinical evaluation.
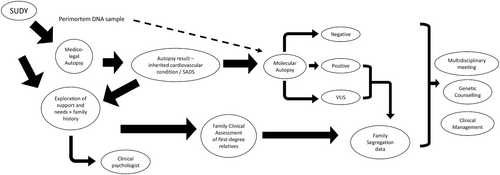
There were around two fifths of family members who had not attended clinical screening, and only one family member was clinically tested positive for an inherited cardiovascular condition. The reports of screening the relatives of SCDY revealed varying yields, ranging from 18% to 30% (Behr et al., 2003; Di Gioia et al., 2006; Kumar et al., 2013; McGorrian et al., 2013; Tan et al., 2005). Several factors contributed to the extremely low yield of clinical screening in our locality, which include a lack of family awareness regarding the potential inheritance of diseases, noncompliance with screening recommendations, long waiting times for public cardiology services, incomplete cardiovascular evaluations, and the incomplete penetrance and variable expressivity of cardiac channelopathy and cardiomyopathy conditions. In fact, the difficulty in arranging cardiological screening for the families is one of the major issues perceived by forensic pathologists in the United Kingdom (Sheppard, 2022). This provides an insight to further support the recommendation that the investigation of SCDY should be overseen by a multidisciplinary team with appropriate expertise in pediatric and/or adult cardiology, genetics, genetic counseling, and pathology (class I recommendation), with familial clinical data obtained fed back to facilitate discussion of segregation data and variant revisiting. (Martínez-Barrios et al., 2023; Stiles et al., 2021). Incorporating the complex needs (medical, psychosocial, spiritual, and financial needs) experienced by parents following SCDY (Ingles et al., 2016; McDonald et al., 2020), a proposed workflow is shown in Figure 2.
4.1 Limitation and further study
The main limitation of the study was its retrospective design, which contributed to incomplete data sets and recall bias. As Chinese families were small in size, therefore, it posed challenges for variant classification due to a lack of cosegregation data in the population. These families were also known to have a high rate of default in follow-up, because these family members were asymptomatic. As a result, variable information from subsequent cascade clinical screening data could potentially be missed. We managed to minimize this bias by contacting the next-of-kin of each families, as part of our follow-up service.
The classification of Chinese ethnicity was based on the history provided by the next-of-kins during forensic examination. There was a possibility of including victims of mixed Chinese and other Asian ethnicities, but this chance should be minimal as Hong Kong is a largely homogenous society with over 90% of the population being Chinese.
VUS were included to be reported in studies of molecular autopsy, because they might potentially explain the causes of young sudden death of decedents, like in previous studies (e.g. Sanchez et al., 2016; Lahrouchi et al., 2017; Hellenthal et al., 2017; Shanks et al., 2018; Scheiper et al., 2018; Larsen et al., 2019, Williams et al., 2019; Iglesias et al., 2021; Ripoll-Vera et al., 2021; Webster et al., 2021; Scheiper-Welling et al., 2022; Rohrer et al., 2023; Wang et al., 2023). The unclear phenotype in gross autopsy-negative victims made it difficult to upgrade these VUS without phenotype and cosegregation data. Revisiting these variants in the future would potentially identify important familial disease causative variants, after availability of clinical screening information of first-degree relatives.
The incidence of SCDY could not be ascertained by the current study as some of the SCDY victims or victims with gross autopsy findings of cardiomyopathy might not have been referred for molecular autopsy due to various reasons. In Hong Kong, all SCDY cases are referred to coroners, who would normally order autopsies when the medical cause of death is not apparent. Since SCDY is sudden and unexpected by definition, the overwhelming majority of these cases would undergo an autopsy examination. In rare instances, coroners can in theory waive autopsies based on nonmedical considerations, particularly a full autopsy investigation is not readily accepted by Asian families. However, such incidents are infrequent. As the molecular autopsy service was solely provided by our single institute in Hong Kong, a good estimation in the number of SCDY victims in the territory could still be reflected.
Furthermore, the study did not cover metabolic and neurological causes of SCDY, which are less common (Chahal et al., 2021; Tsuda et al., 2020).
5 CONCLUSION
The molecular autopsy service enabled the detection of important SCD-causative variants, thereby facilitating family cascade screening. The overall yield of our Chinese cohort was comparable with international cohorts, although there was a higher proportion of VUS compared with LP variants. Our study provided important insights for policy formulation in Asian countries and laid the foundation for a well-structured multidisciplinary program, which aims at supporting SCD victims and families.
AUTHOR CONTRIBUTIONS
Sit-Yee Kwok: Concept or design; acquisition of data; analysis or interpretation of data; drafting of the article. Stephanie Ho: Acquisition of data. Fong-Ying Shih: Acquisition of data; analysis or interpretation of data. Pak-Kwan Yeung: Acquisition of data. Shirley S. W. Cheng: Critical revision for important intellectual content. Wai-Ming Poon: Critical revision for important intellectual content. Ivan F. M Lo: Concept or design; critical revision for important intellectual content. Ho-Ming Luk: Concept or design; acquisition of data; critical revision for important intellectual content. All authors had full access to the data, contributed to the study, approved the final version for publication, and take responsibility for its accuracy and integrity.
ACKNOWLEDGMENTS
We would like to express our gratitude to all the participants involved in this study. Special acknowledgment is given to Dr. Sabrina Tsao, Dr. Jojo Hai, Ms Connie Poon for their support in establishment of the study. Additionally, we also thank Ms Winnie WT Pang in assisting the manuscript management, and Mr. Dick CY Li in providing analytical advice on study data.
FUNDING INFORMATION
Part of the project is supported under Hong Kong Health and Medical Research Grant (HMRF PR-HKCH-1). The funder had no role in study design, data collection/analysis/interpretation or manuscript preparation.
CONFLICT OF INTEREST STATEMENT
All authors have disclosed no conflicts of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.




