New insights into the landscape of ALPL gene variants in patients with hypophosphatasia from the Global HPP Registry
Abstract
Hypophosphatasia (HPP) is a rare, inherited metabolic disease characterized by low tissue-nonspecific alkaline phosphatase activity due to ALPL gene variants. We describe ALPL variants from the observational, prospective, multinational Global HPP Registry. Inclusion in the analysis required a diagnosis of HPP, low serum ALP activity, and ≥1 ALPL variant. Of 1176 patients enrolled as of September 2022, 814 met inclusion criteria in Europe (48.9%), North America (36.7%), Japan (10.2%), Australia (2.6%), and elsewhere (1.6%). Most patients (74.7%) had 1 ALPL variant; 25.3% had ≥2 variants. Nearly all patients (95.6%) had known disease-causing variants; 4.4% had variants of uncertain significance. Disease-causing variants were predominantly missense (770/1556 alleles). The most common variants were c.571G>A (102/1628 alleles), c.1250A>G (66/1628 alleles), and c.1559del (61/1628 alleles). Variant profiles were generally consistent, except in Japan, where a higher proportion of patients (68.7%) had ≥2 ALPL variants, likely because more had disease onset before age 6 months (53.0% vs. 10.1%–23.1% elsewhere). Frameshift mutations (61/164 alleles) and inframe deletions (7/164 alleles) were more common in Japan. Twenty-three novel variants were discovered, each in a single geographic region, predominantly Europe. Analyses confirmed previously known ALPL variants, identified novel variants, and characterized geographic variation in frequency and type of ALPL variants in a large population.
1 INTRODUCTION
Hypophosphatasia (HPP) is a rare, inherited metabolic disorder caused by low tissue-nonspecific alkaline phosphatase (ALP) enzyme activity associated with identifiable loss-of-function variants of the ALPL gene (OMIM 171760) (Johns Hopkins University School of Medicine, 2023; Weiss et al., 1988). Tissue-nonspecific ALP (TNSALP) is a cell-surface phosphohydrolase expressed in the bone, kidney, and liver (Weiss et al., 1988). Deficient activity of ALP can lead to extracellular accumulation of its substrates inorganic pyrophosphate (PPi) and pyridoxal 5′-phosphate (PLP) (Whyte, 2010). The accumulation of PPi can cause defective mineralization of bones and teeth and pseudogout (Whyte, 2010; Whyte, 2016). Impaired hydrolysis of PLP can cause vitamin B6 deficiency in the central nervous system and reduced neurotransmitter synthesis, potentially leading to pyridoxine-responsive seizures in severely affected infants (Baumgartner-Sigl et al., 2007; Whyte, 2016). A third TNSALP substrate and biochemical marker for HPP, phosphoethanolamine (PEA), is elevated in patients with HPP but has unclear physiologic significance. PEA levels can be obtained by ordering plasma or urine amino acids testing (Whyte, 2016).
When the most severe form of HPP manifests, generally during the perinatal and infantile stages of life (before age 6 months), it is associated with significant mortality, vitamin B6–responsive seizures, respiratory failure, and failure to thrive (Whyte, 2016). Other manifestations across patients of different ages include premature loss of deciduous teeth in children, bone deformity, rickets (in children prior to growth plate closure), osteomalacia, pseudofractures, recurrent and poorly healing fractures (e.g., metatarsal fractures, atypical femoral fractures), fatigue, muscle weakness, bone pain, difficulty walking, and reduced quality of life (Dahir et al., 2022; Dahir et al., 2023; Genest & Seefried, 2018; Högler et al., 2019; Rauf et al., 2023; Szabo et al., 2019).
HPP follows either an autosomal recessive mode of inheritance with homozygous or compound heterozygous ALPL variants or an autosomal dominant mode of inheritance with heterozygous, usually dominant-negative ALPL variants (Hancarova et al., 2015; Linglart & Biosse-Duplan, 2016; Mornet et al., 2021; Taillandier et al., 2018; Watanabe et al., 2014). However, genotype–phenotype correlation is poor in HPP. Although HPP that manifests before 6 months of age may be considered to have less phenotypic variability than HPP that manifests after 6 months of age, some variability in disease signs and symptoms was reported in the Mennonite perinatal/infantile HPP population (Leung et al., 2013). Furthermore, prenatal benign HPP, which is characterized by prenatal findings suggestive of severe bone dysplasia but a benign postnatal course, can have autosomal dominant inheritance with apparent maternal bias of transmission (Moore et al., 1999; Pauli et al., 1999). Phenotypic variability is broader in pediatric and adult populations, both within and between families, most often with dominant variants but with autosomal recessive variants as well (Henthorn et al., 1992; Hofmann et al., 2014; Kato et al., 2020; Mornet, 2018). Thus, other genetic factors, as well as environmental and epigenetic factors, likely contribute to the varying clinical manifestations of HPP (Hofmann et al., 2014; Mornet et al., 2021).
Genetic testing is not mandatory at the perinatal/infantile end of the HPP disease spectrum because of the severity of the signs and symptoms. However, sequencing of the ALPL gene (chromosome 1p36.1-p34) is recommended for definitive diagnosis (Bianchi, 2015; Michigami, Ohata, et al., 2020; Simon et al., 2018). To date, more than 440 ALPL variants (including missense, nonsense, splice site, and frameshift deletions and insertions) have been described in the Johannes Kepler University (JKU) ALPL Gene Variant Database (Farman et al., 2024; JKU Faculty of Medicine, 2024). A variety of sequencing techniques have been used to detect these variants, including single-gene, panel, targeted, whole-exome, and whole-genome sequencing (De Sousa et al., 2018; Kishnani et al., 2017; Taillandier et al., 2015). However, ALPL variants may not always be detected by standard sequencing techniques, although they do capture an estimated 95% of the variants in patients with HPP (Mornet et al., 2014). False-negative genetic results may occur in cases of intronic mutations or deletions/duplications or for other reasons, including the type of test used (De Sousa et al., 2018). Additionally, variants of uncertain significance (VUS) may go unreported by some laboratories (Beck et al., 2023). Given insufficient evidence for a VUS to be causative of HPP, additional assessment through variant reclassification of the VUS is warranted and can be performed through the ALPL Gene Variant Consortium, with all variants, genotypes, and phenotypes displayed in the JKU ALPL Gene Variant Database (Farman et al., 2024; JKU Faculty of Medicine, 2024). Ultimately, a negative ALPL genetic test result thus does not rule out HPP, although careful phenotyping and assessment of biomarkers are essential to exclude a multitude of other possible causes of low ALP activity (Beck et al., 2023; Riancho-Zarrabeitia et al., 2016; Saraff et al., 2016). HPP is a clinical diagnosis for which genotyping is not required to confirm (Khan et al., 2019; Khan et al., 2024).
The Global HPP Registry, the largest observational study of patients with HPP to date, was designed to collect data about the natural history of HPP and the long-term effectiveness and safety of treatment with asfotase alfa (Högler et al., 2019; Seefried et al., 2020). Genetic data are also being collected in the registry, allowing for the characterization of the range of ALPL variants associated with HPP and their geographic distribution. The current investigation is a foundational report and the first of a series of planned analyses of ALPL variant data from the Global HPP Registry. While future reports will focus on genotype–phenotype correlations, the present analysis focuses on the geographic distribution and frequencies of ALPL variants, including the novel, not yet reported variants.
2 METHODS
2.1 Editorial policies and ethical considerations
The study protocol for the Global HPP Registry was approved by the institutional review board (or local equivalent) of participating study sites. Data collection and analysis was conducted in accordance with International Conference on Harmonisation Good Clinical Practice Guidelines and the Declaration of Helsinki. All participants provided written informed consent prior to enrollment.
2.2 Study design
The Global HPP Registry is an observational, prospective, multinational study (NCT02306720; EUPAS13514) initiated in 2014 to record data from patients diagnosed with HPP regardless of asfotase alfa treatment status (Högler et al., 2019). Study design and data collection have been described (Dahir et al., 2022; Dahir et al., 2023; Högler et al., 2019; Seefried et al., 2020). Registry data used in these analyses were collected from initiation through September 5, 2022.
2.3 Patient eligibility
Patients of any age meeting the following criteria related to data reported to the Global HPP Registry were eligible for inclusion: diagnosis of HPP, documented registry enrollment date, serum ALP activity lower than the age- and sex-adjusted reference range, and genetic testing results reporting any ALPL variant designated as disease-causing or as a VUS. Variants were designated as disease-causing by submitters to the Global HPP Registry if they met the criteria for a pathogenic (P) or likely pathogenic (LP) classification under the American College of Medical Genetics and Genomics (ACMG) guidelines (Richards et al., 2015).
2.4 Data collection and classification
All ALPL variants reported in the registry were reviewed, and ambiguous or incomplete data were clarified through requests to individual sites and consultations with expert geneticists. Most variant classifications were taken directly from the Global HPP Registry entries. However, the registry contained conflicting submissions and infrequent missing entries, which were resolved by referencing the VarSome and ClinVar databases and the ALPL Gene Variant Database. In cases where variant classifications conflicted between registry entries, variants were categorized according to the ACMG classification reported in these databases; if there was no conflict, the original registry submission classification was used. If there was a conflict between VarSome, ClinVar and the ALPL Gene Variant Database, the classification reported in the majority of databases was used. Resolution of the pathogenicity classifications of conflicting registry submissions is summarized in Table S1.
Benign (B) and likely benign (LB) variants were not considered in the current analysis. Variants were categorized as novel if they differed from all existing variants on the canonical ALPL transcript reported in Ensembl's supporting databases (including gnomAD and ClinVar), HGMD Pro, and ALPL Gene Variant Database. Functional effects of all variants were predicted using Ensembl Variant Effect Predictor with respect to the canonical ALPL transcript (ENST00000374840.8). The mode of inheritance in patients with two different ALPL variants was classified as putative compound heterozygous because inheritance in trans could not be established with parental studies and after attempts to confirm they were not haplotypes. Frequently recurrent variant pairs were evaluated for their presence as frequent haplotypes in the large-scale biomedical database and research resource UK Biobank. Perinatal/infantile HPP was defined as disease onset at age ≤6 months.
2.5 Data analysis
3 RESULTS
3.1 Study population
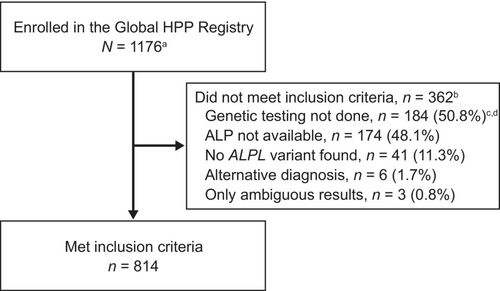
Of 1176 patients enrolled in the Global HPP Registry, 814 met inclusion criteria (Figure 1). Among the 814 patients, 63.9% were female and 71.9% were White (Table 1). Patients were from Europe (48.9%), North America (36.7%), Japan (10.2%), Australia (2.6%), and other regions (1.6%). A majority (68.6%) of the population had HPP onset at age ≥6 months, ranging from 43.4% to 76.9% across regions (Table 2). The frequency of perinatal/infantile HPP was 53.0% in Japan compared with 10.1%–23.1% outside of Japan. In the study population, 41.0% of patients were ever treated with asfotase alfa enzyme replacement therapy.
| Patients N = 814 | |
|---|---|
| Age at diagnosis of HPP, y | n = 740 |
| Median (IQR) | 13.7 (2.5–43.5) |
| Sex, n (%) | n = 814 |
| Female | 520 (63.9) |
| Race, n (%) | n = 814 |
| White | 585 (71.9) |
| Asian | 99 (12.2) |
| Native Hawaiian or other Pacific Islander | 2 (0.2) |
| Native American or Alaska Native | 1 (0.1) |
| Black or African American | 1 (0.1) |
| Other/multiple | 21 (2.6) |
| Unknown | 105 (12.9) |
| Ethnicity, n (%) | n = 814 |
| Not Hispanic or Latino | 628 (77.1) |
| Hispanic or Latino | 29 (3.6) |
| Unknown | 157 (19.3) |
| Geographic region, n (%) | n = 814 |
| Europea | 398 (48.9) |
| North Americab | 299 (36.7) |
| Japan | 83 (10.2) |
| Australia | 21 (2.6) |
| Otherc | 13 (1.6) |
- Abbreviations: HPP, hypophosphatasia; IQR, interquartile range.
- a Includes Austria, Belgium, France, Germany, Greece, Ireland, Italy, Poland, Portugal, Spain, and United Kingdom.
- b Includes Canada and United States.
- c Includes Israel, Russia, Saudi Arabia, Taiwan, and Turkey.
| All N = 814 | Europea n = 398 | North Americab n = 299 | Japan n = 83 | Australia n = 21 | Otherc n = 13 | |
|---|---|---|---|---|---|---|
| Age of HPP onset, n (%) | ||||||
| <6 months | 122 (15.0) | 40 (10.1) | 32 (10.7) | 44 (53.0) | 3 (14.3) | 3 (23.1) |
| ≥6 months | 558 (68.6) | 276 (69.3) | 220 (73.6) | 36 (43.4) | 16 (76.2) | 10 (76.9) |
| Unknown/unspecified | 134 (16.5) | 82 (20.6) | 47 (15.7) | 3 (3.6) | 2 (9.5) | 0 |
| Number of ALPL variants, n (%) | ||||||
| 1 | 608 (74.7) | 301 (75.6) | 253 (84.6) | 26 (31.3) | 18 (85.7) | 10 (76.9) |
| ≥2d | 206 (25.3) | 97 (24.4) | 46 (15.4) | 57 (68.7) | 3 (14.3) | 3 (23.1) |
| Variant type, n (%) | ||||||
| Disease-causing (P or LP)e | 778 (95.6) | 382 (96.0) | 285 (95.3) | 82 (98.8) | 20 (95.2) | 9 (69.2) |
| VUS only | 36 (4.4) | 16 (4.0) | 14 (4.7) | 1 (1.2) | 1 (4.8) | 4 (30.8) |
| ≥1 VUS, n (%) | 51 (6.3) | 22 (5.5) | 21 (7.0) | 3 (3.6) | 1 (4.8) | 4 (30.8) |
| Allele count/total alleles (VAF %) | ||||||
| Disease-causing alleles | 965/1628 (59.3) | 470/796 (59.0) | 324/598 (54.2) | 136/166 (81.9) | 23/42 (54.8) | 12/26 (46.2) |
| VUS alleles | 56/1628 (3.4) | 26/796 (3.3) | 21/598 (3.5) | 4/166 (2.4) | 1/42 (2.4) | 4/26 (15.4) |
- Abbreviations: HPP, hypophosphatasia; LP, likely pathogenic; P, pathogenic; VAF, variant allele frequency; VUS, variant of uncertain significance.
- a Includes Austria, Belgium, France, Germany, Greece, Ireland, Italy, Poland, Portugal, Spain, and United Kingdom.
- b Includes Canada and United States.
- c Includes Israel, Russia, Saudi Arabia, Taiwan, and Turkey.
- d One patient had 3 ALPL variants, and 204 patients had 2 ALPL variants.
- e Some patients with disease-causing variants also had VUS.
Among 797 patients with available data on the gene sequencing method used, 399 (50.1%) had single-gene sequencing, 108 (13.6%) had targeted sequencing, 51 (6.4%) had whole-exome sequencing, and 33 (4.1%) had panel sequencing; the sequencing method was unknown or other for the remaining 206 patients (25.9%).
3.2 ALPL genotypes in the overall population
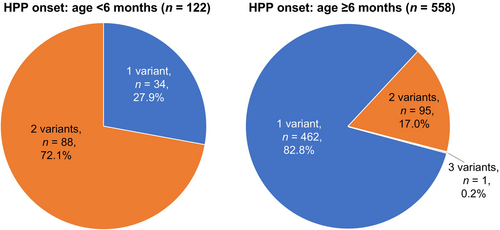
In total, 1021 variant ALPL alleles representing 252 unique sequence variants were identified in 814 patients. The distribution of ALPL variants is shown across the gene sequence in Figure 2a and across the TNSALP protein structure in Figure 2b; no hot spots were apparent in reference to previously described TNSALP protein domains (Silvent et al., 2014; Yu et al., 2023); domain ranges are provided in Table S2. Three quarters of patients (74.7% [608/814]) had a single ALPL variant, 25.2% (205/814) had two variants, and 0.1% (1/814) had three variants (Table 2). Among patients with a single ALPL variant, 164 had a variant that exerts a dominant-negative effect according to in vitro testing (del Angel et al., 2020) (e.g., c.1250A>G; 4.7% of patients) and 227 had a variant with no dominant-negative effect (eg, c.571G>A; 3.0% of patients). The remaining 217 patients had a variant without dominant-negative testing results. The presence of three variants in one patient indicates that at least two of these variants were located on the same allele, although phasing data were unavailable to evaluate which variants were in cis. Multiple ALPL variants occurred more frequently in patients with disease onset at age ≤6 months (72.1% [88/122]) than in those with disease onset at age ≥6 months or unknown (17.2% [96/558]; chi-square p < 0.0001; Figure 3). Among 206 patients in whom two or more variants were identified, 173 (84.0%) had putative compound heterozygous variants and 33 (16.0%) had homozygous variants. Patients with HPP onset at age ≤6 months had a higher proportion of compound heterozygous (69/122 [56.6%]) and a similar proportion of homozygous (19/122 [15.6%]) variants compared with patients with a later or unspecified age of disease onset (compound heterozygous: 83/558 [14.9%]; homozygous: 13/558 [2.3%]).
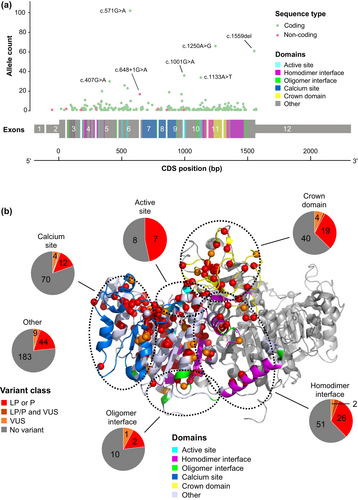
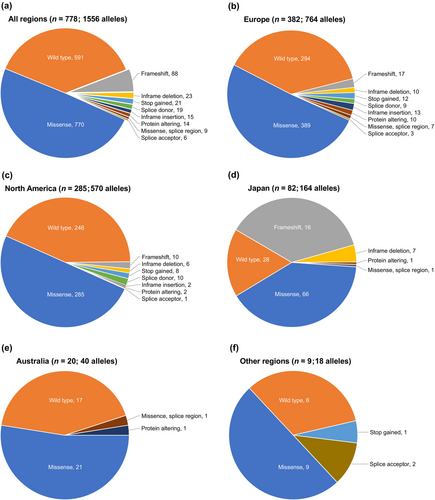
Most patients (778 [95.6%]) had at least one disease-causing (P or LP) variant, whereas 36 patients (4.4%) had only VUS (Table 2). The most common disease-causing variants were missense (770/1556 alleles) and frameshift variants (88/1556 alleles) (Figure 4a). Most VUS were missense (45/102 alleles).
Heterozygous genotypes that were observed in ≥3% of patients globally were c.1250A>G (7.0%), c.571G>A (4.4%), and c.1133A>T (3.9%; Table 3). The two most frequent homozygous genotypes were c.1559del/c.1559del (1.4%) and c.571G>A/c.571G>A (0.6%). A total of 123 unique pairs of putative compound heterozygous variants (including pairs with large deletions) were reported, 25 of which were reported two or more times. Only one of these recurring pairs (c.571G>A/c.881A>C) was found in the UK Biobank (UK Biobank, 2023), but the frequency of this pair was too low to support the presence of a haplotype (D′ = 0.006). The two most frequent putative compound heterozygous genotypes were c.1001G>A/c.571G>A (13 patients [1.6%]) and c.1559del/c.979T>C (9 patients [1.1%]).
| Genotype, n (%) of patients | All n = 814 | Europea n = 398 | North Americab n = 299 | Japan n = 83 | Australia n = 21 | Otherc n = 13 |
|---|---|---|---|---|---|---|
| Heterozygous genotypes reported in ≥2.0% of patients in any region and >1 patient overall | ||||||
| c.1250A>G | 57 (7.0) | 10 (2.5) | 38 (12.7) | 0 | 9 (42.9) | 0 |
| c.571G>A | 36 (4.4) | 18 (4.5) | 15 (5.0) | 0 | 0 | 3 (23.1) |
| c.1133A>T | 32 (3.9) | 2 (0.5) | 30 (10.0) | 0 | 0 | 0 |
| c.1001G>A | 19 (2.3) | 9 (2.3) | 10 (3.3) | 0 | 0 | 0 |
| c.550C>T | 18 (2.2) | 9 (2.3) | 4 (1.3) | 5 (6.0) | 0 | 0 |
| c.346G>A | 16 (2.0) | 6 (1.5) | 10 (3.3) | 0 | 0 | 0 |
| c.407G>A | 16 (2.0) | 12 (3.0) | 3 (1.0) | 1 (1.2) | 0 | 0 |
| c.881A>C | 14 (1.7) | 4 (1.0) | 9 (3.0) | 0 | 1 (4.8) | 0 |
| c.1559del | 11 (1.4) | 0 | 0 | 11 (13.3) | 0 | 0 |
| c.212G>A | 10 (1.2) | 5 (1.3) | 4 (1.3) | 0 | 1 (4.8) | 0 |
| c.343_348dup | 8 (1.0) | 8 (2.0) | 0 | 0 | 0 | 0 |
| c.1426G>A | 8 (1.0) | 6 (1.5) | 1 (0.3) | 0 | 1 (4.8) | 0 |
| c.526G>A | 8 (1.0) | 3 (0.8) | 4 (1.3) | 0 | 1 (4.8) | 0 |
| c.575T>C | 8 (1.0) | 1 (0.3) | 7 (2.3) | 0 | 0 | 0 |
| c.1144G>A | 6 (0.7) | 1 (0.3) | 3 (1.0) | 2 (2.4) | 0 | 0 |
| c.1283G>A | 6 (0.7) | 3 (0.8) | 1 (0.3) | 0 | 2 (9.5) | 0 |
| c.871G>A | 4 (0.5) | 3 (0.8) | 0 | 0 | 0 | 1 (7.7) |
| c.984_986del | 4 (0.5) | 1 (0.3) | 1 (0.3) | 2 (2.4) | 0 | 0 |
| c.203C>T | 3 (0.4) | 2 (0.5) | 0 | 0 | 1 (4.8) | 0 |
| c.299C>T | 2 (0.2) | 1 (0.3) | 0 | 0 | 1 (4.8) | 0 |
| c.298-1G>C | 2 (0.2) | 0 | 0 | 0 | 0 | 2 (15.4) |
| Putative compound heterozygousd genotypes reported in >1 patient | ||||||
| c.1001G>A/c.571G>A | 13 (1.6) | 7 (1.8) | 6 (2.0) | 0 | 0 | 0 |
| c.1559del/c.979T>C | 9 (1.1) | 0 | 0 | 9 (10.8) | 0 | 0 |
| c.1250A>G/c.571G>A | 4 (0.5) | 3 (0.8) | 1 (0.3) | 0 | 0 | 0 |
| c.1130C>T/c.1559del | 3 (0.4) | 0 | 0 | 3 (3.6) | 0 | 0 |
| c.346G>A/c.571G>A | 3 (0.4) | 1 (0.3) | 2 (0.7) | 0 | 0 | 0 |
| c.407G>A/c.512A>G | 3 (0.4) | 3 (0.8) | 0 | 0 | 0 | 0 |
| c.526G>A/c.648+1G>A | 3 (0.4) | 2 (0.5) | 1 (0.3) | 0 | 0 | 0 |
| Deletion of exon 12 (3′ part)/ c.648+1G>A | 2 (0.2) | 2 (0.5) | 0 | 0 | 0 | 0 |
| c.1024_1025delGAinsTT/c.1195G>T | 2 (0.2) | 0 | 2 (0.7) | 0 | 0 | 0 |
| c.1130C>T/c.571G>A | 2 (0.2) | 2 (0.5) | 0 | 0 | 0 | 0 |
| c.113_116del/c.118dup | 2 (0.2) | 2 (0.5) | 0 | 0 | 0 | 0 |
| c.1144G>A/c.529G>A | 2 (0.2) | 0 | 0 | 2 (2.4) | 0 | 0 |
| c.1171C>T/c.571G>A | 2 (0.2) | 2 (0.5) | 0 | 0 | 0 | 0 |
| c.1183A>G/c.1559del | 2 (0.2) | 0 | 0 | 2 (2.4) | 0 | 0 |
| c.1318_1320del/c.571G>A | 2 (0.2) | 1 (0.3) | 1 (0.3) | 0 | 0 | 0 |
| c.1348C>T/c.1559del | 2 (0.2) | 0 | 0 | 2 (2.4) | 0 | 0 |
| c.1559del/c.407G>A | 2 (0.2) | 0 | 0 | 2 (2.4) | 0 | 0 |
| c.1559del/c.572A>G | 2 (0.2) | 0 | 0 | 2 (2.4) | 0 | 0 |
| c.211C>T/c.571G>A | 2 (0.2) | 2 (0.5) | 0 | 0 | 0 | 0 |
| c.227A>G/c.571G>A | 2 (0.2) | 2 (0.5) | 0 | 0 | 0 | 0 |
| c.406C>T/c.979T>C | 2 (0.2) | 0 | 0 | 2 (2.4) | 0 | 0 |
| c.500C>T/c.571G>A | 2 (0.2) | 2 (0.5) | 0 | 0 | 0 | 0 |
| c.550C>T/c.571G>A | 2 (0.2) | 0 | 2 (0.7) | 0 | 0 | 0 |
| c.571G>A/c.881A>C | 2 (0.2) | 1 (0.3) | 1 (0.3) | 0 | 0 | 0 |
| Homozygous genotypes reported in >1 patient | ||||||
| c.1559del/c.1559del | 11 (1.4) | 0 | 0 | 11 (13.3) | 0 | 0 |
| c.571G>A/c.571G>A | 5 (0.6) | 2 (0.5) | 3 (1.0) | 0 | 0 | 0 |
| c.715G>T/c.715G>T | 3 (0.4) | 3 (0.8) | 0 | 0 | 0 | 0 |
| c.293C>T/c.293C>T | 2 (0.2) | 0 | 0 | 0 | 0 | 2 (15.4) |
- a Includes Austria, Belgium, France, Germany, Greece, Ireland, Italy, Poland, Portugal, Spain, and United Kingdom.
- b Includes Canada and United States.
- c Includes Israel, Russia, Saudi Arabia, Taiwan, and Turkey.
- d Refers to presence of ≥2 variants; this analysis did not differentiate whether variants were on 1 or 2 alleles.
Globally, the three most common disease-causing variants were c.571G>A (VAF: 6.3% [102/1628 alleles]), c.1250A>G (4.1% [66/1628 alleles]), and c.1559del (3.7% [61/1628 alleles]; Table 4). The most common VUS were c.715G>T (0.5% [8/1628 alleles]) and c.1156G>T (0.2% [4/1628 alleles]). A total of 23 unique novel ALPL variants were identified in 30 patients (30 alleles; Table 5).
| Allele count (VAF %)a | All n = 814 patients, 1628 alleles | Europeb n = 398 patients, 796 alleles | North Americac n = 299 patients, 598 alleles | Japan n = 83 patients, 166 alleles | Australia n = 21 patients, 42 alleles | Otherd n = 13 patients, 26 alleles |
|---|---|---|---|---|---|---|
| P/LP variants reported in ≥2.0% of alleles in any region and >1 patient overall | ||||||
| c.571G>A | 102 (6.3) | 59 (7.4) | 39 (6.5) | 0 | 0 | 4 (15.4) |
| c.1250A>G | 66 (4.1) | 15 (1.9) | 42 (7.0) | 0 | 9 (21.4) | 0 |
| c.1559del | 61 (3.7) | 0 | 0 | 61 (36.7) | 0 | 0 |
| c.1001G>A | 36 (2.2) | 17 (2.1) | 19 (3.2) | 0 | 0 | 0 |
| c.1133A>T | 34 (2.1) | 2 (0.3) | 32 (5.4) | 0 | 0 | 0 |
| c.407G>A | 30 (1.8) | 22 (2.8) | 4 (0.7) | 3 (1.8) | 1 (2.4) | 0 |
| c.526G>A | 26 (1.6) | 12 (1.5) | 12 (2.0) | 0 | 2 (4.8) | 0 |
| c.550C>T | 23 (1.4) | 10 (1.3) | 6 (1.0) | 7 (4.2) | 0 | 0 |
| c.346G>A | 20 (1.2) | 7 (0.9) | 13 (2.2) | 0 | 0 | 0 |
| c.881A>C | 19 (1.2) | 6 (0.8) | 12 (2.0) | 0 | 1 (2.4) | 0 |
| c.648+1G>A | 17 (1.0) | 8 (1.0) | 9 (1.5) | 0 | 0 | 0 |
| c.979T>C | 17 (1.0) | 0 | 0 | 17 (10.2) | 0 | 0 |
| c.1171C>T | 11 (0.7) | 4 (0.5) | 6 (1.0) | 1 (0.6) | 0 | 0 |
| c.212G>A | 11 (0.7) | 6 (0.8) | 4 (0.7) | 0 | 1 (2.4) | 0 |
| c.984_986del | 10 (0.6) | 3 (0.4) | 2 (0.3) | 5 (3.0) | 0 | 0 |
| c.343_348dup | 10 (0.6) | 10 (1.3) | 0 | 0 | 0 | 0 |
| c.1144G>A | 8 (0.5) | 1 (0.1) | 3 (0.5) | 4 (2.4) | 0 | 0 |
| c.1426G>A | 8 (0.5) | 6 (0.8) | 1 (0.2) | 0 | 1 (2.4) | 0 |
| c.318G>C | 7 (0.4) | 3 (0.4) | 3 (0.5) | 0 | 1 (2.4) | 0 |
| c.1283G>A | 6 (0.4) | 3 (0.4) | 1 (0.2) | 0 | 2 (4.8) | 0 |
| c.871G>A | 4 (0.2) | 3 (0.4) | 0 | 0 | 0 | 1 (3.8) |
| c.293C>T | 4 (0.2) | 0 | 0 | 0 | 0 | 4 (15.4) |
| c.203C>T | 3 (0.2) | 2 (0.3) | 0 | 0 | 1 (2.4) | 0 |
| Deletion of exon 9 (3′ part) | 3 (0.2) | 2 (0.3) | 0 | 0 | 1 (2.4) | 0 |
| c.299C>T | 3 (0.2) | 2 (0.3) | 0 | 0 | 1 (2.4) | 0 |
| c.303C>A | 3 (0.2) | 1 (0.1) | 1 (0.2) | 0 | 0 | 1 (3.8) |
| c.298-1G>C | 2 (0.1) | 0 | 0 | 0 | 0 | 2 (7.7) |
| VUS reported in >1 patient overall | ||||||
| c.715G>T | 8 (0.5) | 8 (1.0) | 0 | 0 | 0 | 0 |
| c.1156G>T | 4 (0.2) | 0 | 4 (0.7) | 0 | 0 | 0 |
| c.1034C>T | 3 (0.2) | 0 | 3 (0.5) | 0 | 0 | 0 |
| c.818C>T | 3 (0.2) | 0 | 2 (0.3) | 0 | 0 | 1 (3.8) |
| c.1573T>C | 2 (0.1) | 2 (0.3) | 0 | 0 | 0 | 0 |
| c.262G>A | 2 (0.1) | 2 (0.3) | 0 | 0 | 0 | 0 |
| c.436G>A | 2 (0.1) | 1 (0.1) | 1 (0.2) | 0 | 0 | 0 |
- Abbreviations: LP, likely pathogenic; P, pathogenic; VAF, variant allele frequency; VUS, variant of uncertain significance.
- a Allele count is defined as the number of alleles on which the variant was observed, and % indicates the percentage of all alleles (2× no. of patients) with the indicated variant.
- b Includes Austria, Belgium, France, Germany, Greece, Ireland, Italy, Poland, Portugal, Spain, and United Kingdom.
- c Includes Canada and United States.
- d Includes Israel, Russia, Saudi Arabia, Taiwan, and Turkey.
| All n = 814 | Europea n = 398 | North Americab n = 299 | Japan n = 83 | Australia n = 21 | Otherc n = 13 | |
|---|---|---|---|---|---|---|
| Patients with novel variants, n (%) | 30 (3.7) | 18 (4.5) | 9 (3.0) | 3 (3.6) | 0 | 0 |
| No. of novel variant alleles | 30 | 18 | 9 | 3 | 0 | 0 |
| No. of unique novel variants | 23 | 14 | 6 | 3 | 0 | 0 |
| Unique novel variants, n (% of total unique novel variants) | ||||||
| c.1018C>T | 3 (13.0) | 3 (21.4) | 0 | 0 | 0 | 0 |
| c.1327G>T | 3 (13.0) | 0 | 3 (50.0) | 0 | 0 | 0 |
| c.1024_1025delGAinsTT | 2 (8.7) | 0 | 2 (33.3) | 0 | 0 | 0 |
| c.1033G>C | 2 (8.7) | 2 (14.3) | 0 | 0 | 0 | 0 |
| c.118dup | 2 (8.7) | 2 (14.3) | 0 | 0 | 0 | 0 |
| c.1043C>A | 1 (4.3) | 0 | 1 (16.7) | 0 | 0 | 0 |
| c.1156G>C | 1 (4.3) | 1 (7.1) | 0 | 0 | 0 | 0 |
| c.1207A>C | 1 (4.3) | 1 (7.1) | 0 | 0 | 0 | 0 |
| c.1213A>C | 1 (4.3) | 0 | 1 (16.7) | 0 | 0 | 0 |
| c.1252G>A | 1 (4.3) | 1 (7.1) | 0 | 0 | 0 | 0 |
| c.1356G>A | 1 (4.3) | 0 | 0 | 1 (33.3) | 0 | 0 |
| c.1356G>T | 1 (4.3) | 1 (7.1) | 0 | 0 | 0 | 0 |
| c.1451T>C | 1 (4.3) | 1 (7.1) | 0 | 0 | 0 | 0 |
| c.1482_1532del | 1 (4.3) | 0 | 0 | 1 (33.3) | 0 | 0 |
| c.1491_1492del | 1 (4.3) | 1 (7.1) | 0 | 0 | 0 | 0 |
| c.182-1G>A | 1 (4.3) | 1 (7.1) | 0 | 0 | 0 | 0 |
| c.206C>T | 1 (4.3) | 1 (7.1) | 0 | 0 | 0 | 0 |
| c.304_312dup | 1 (4.3) | 0 | 1 (16.7) | 0 | 0 | 0 |
| c.364_365del | 1 (4.3) | 0 | 1 (16.7) | 0 | 0 | 0 |
| c.380C>T | 1 (4.3) | 1 (7.1) | 0 | 0 | 0 | 0 |
| c.560A>G | 1 (4.3) | 1 (7.1) | 0 | 0 | 0 | 0 |
| c.955A>T | 1 (4.3) | 1 (7.1) | 0 | 0 | 0 | 0 |
| c.981C>G | 1 (4.3) | 0 | 0 | 1 (33.3) | 0 | 0 |
- a Includes Austria, Belgium, France, Germany, Greece, Ireland, Italy, Poland, Portugal, Spain, and United Kingdom.
- b Includes Canada and United States.
- c Includes Israel, Russia, Saudi Arabia, Taiwan, and Turkey.
3.3 Regional variation in ALPL genotype
Consistent with the overall population, most patients in each geographic region had a single ALPL variant (ranging from 76.9% [10/13] in the group named “other regions” to 85.7% [18/21] in Australia), except in Japan, where only 31.3% (26/83) of patients had a single ALPL variant and 68.7% (57/83) had two or more ALPL variants (Table 2). In general, patients with two or more variants were most commonly putatively compound heterozygous in each region: Europe (88.7% [86/97]), North America (89.1% [41/46]), Japan (75.4% [43/57]), and Australia (66.7% [2/3]). At least 95% of patients in Europe, North America, Australia, and Japan had known disease-causing variants; the proportion of patients with only VUS ranged from 1.2% (1/83) in Japan to 4.8% (1/21) in Australia (Table 2).
Predicted effects of disease-causing variants were generally consistent across regions outside of Japan, with missense variants identified in 50.9% of alleles (389/764) in Europe to 52.5% (21/40) in Australia, frameshift variants identified in up to 2.2% of alleles (17/764) in Europe, and inframe deletions identified in up to 1.3% of alleles (10/764) in Europe (Figure 4b–f). The Japanese population had a lower proportion of missense alleles (40.2% [66/164]) and a higher proportion of frameshift (37.2% [61/164]) or inframe deletions (4.3% [7/164]) compared with all other regions (Figure 4d).
The most common variant overall, c.571G>A, was in the top four most frequent variants in Europe, North America, and other regions, and the second most common variant overall, c.1250A>G, was in the top four in Europe, North America, and Australia (Table 4). The variants c.1250A>G and c.1133A>T were more common in North America (7.0% [42/598 alleles] and 5.4% [32/598 alleles], respectively) than in Europe (1.9% [15/796 alleles] and 0.3% [2/796 alleles], respectively). Several common variants (e.g., c.571G>A, c.1250A>G, c.1001G>A, c.1133A>T) were not reported in the Japanese population (Table 4). By contrast, the most common variants in Japan, c.1559del (36.7% [61/166 alleles]) and c.979T>C (10.2% [17/166 alleles]), were reported exclusively in Japan, where 75.0% (33/44) of patients with perinatal/infantile-onset HPP had c.1559del compared with 43.6% (17/39) of patients with HPP onset after age 6 months or with an unspecified age of onset.
The prevalence of VUS differed between geographic regions (Table 4), with c.715G>T reported only in Europe and c.1156G>T and c.1034C>T reported only in North America and none of these reported in Japan.
Of the 23 unique novel variants (Table S3), 14 (60.9%) were reported in Europe, 6 (26.1%) in North America, and 3 (13.0%) in Japan (Table 5). Each novel variant was reported only in a single geographic region. Most patients with novel variants were White/Caucasian (70% [21/30 patients]); 20.0% [6/30] were Asian, and 10.0% [3/30] had unreported race.
4 DISCUSSION
We describe the genetic characteristics of a global cohort of 814 patients with HPP enrolled in the Global HPP Registry since its establishment in 2014. A total of 252 unique ALPL variants were identified in the population analyzed. Three quarters of the patients had a single ALPL variant, and 25% had two ALPL variants. Nearly all variant alleles were previously reported disease-causing variants, and most were missense, a finding that aligns with the set of known variants (JKU Faculty of Medicine, 2024).
Variant number per patient, predicted functional consequences, and allele frequencies varied slightly between Europe and North America and were remarkably distinct in Japan. The three most frequent variants have been previously linked to founder effects in the European (c.571G>A and c.1250A>G) and Japanese (c.1559del) populations (Michigami et al., 2005; Taketani et al., 2014). Geographic variation was also observed in the most prevalent VUS, with c.1156G>T and c.1034C>T reported exclusively in North America, c.715G>T reported only in Europe, and none of these three variants reported in Japan.
Globally, patients with disease onset at age ≤6 months were more likely to have two or more ALPL variants than those with later HPP onset, although compound heterozygosity could not be verified in patients with multiple variants because of limitations in data collection. Frequently observed variant pairs in populations in Europe (c.1001G>A/c.571G>A) and Japan (c.1559del/c.979T>C) have been previously associated with high phenotypic variability (Hofmann et al., 2014; Michigami, Tachikawa, et al., 2020). Additionally, 23 novel variants were discovered in the Global HPP Registry in the current analysis, with most reported only in Europe.
The relative frequencies and geographic distribution of disease-causing variants in the Global HPP Registry are broadly consistent with those observed in large population databases. The most frequent pathogenic variants found among registry cases in Europe (c.571G>A and c.1250A>G) and Japan (c.1559del) are also frequently found in gnomAD in European and East Asian populations, as well as other clinical databases such as ClinVar and the ALPL Gene Variant Database (Farman et al., 2024; Karczewski et al., 2019; Karczewski et al., 2020). As expected, the Global HPP Registry has a higher proportion of these frequent disease-causing variants relative to the general population. For example, the variants c.571G>A and c.1250A>G are 78 times (7.4% vs 0.095%) and 475 times (1.9% vs. 0.004%) more prevalent, respectively, among Europeans in the Global HPP Registry than in gnomAD 4.0 among non-Finnish European individuals. Monoallelic cases show greater enrichment for variants with evidence of dominant-negative effects, such as c.1250A>G (408 times more prevalent; 1.7% vs. 0.004% in Europeans) and c.1001G>A (8312 times more prevalent; 1.5% vs. 0.00018% in Europeans), than for non–dominant-negative variants, such as c.571G>A (32 times more prevalent; 3.0% vs. 0.095% in Europeans). This suggests a comparatively low penetrance of non–dominant-negative variants such as c.571G>A in the monoallelic, heterozygous state. However, it should be noted that variants without dominant-negative effects constituted a substantial fraction of monoallelic cases in the registry. Of 608 monoallelic patients, 164 had variants with documented dominant-negative effects (del Angel et al., 2020), 227 had variants without dominant-negative effects, and the remaining 217 had untested variants. The unknown penetrance and spectrum of disease among individuals heterozygous for disease-causing variants remain important issues that future studies of registry genotype–phenotype correlation may help to resolve.
Notably, prevalent variants of uncertain significance in the Global HPP Registry also showed substantial enrichment over population databases. The most common VUS in Europe, c.715G>T (5 patients, 8 alleles) was found in 1% of European registry patients but was not found in 556,005 non-Finnish Europeans in gnomAD 4.0. This variant has conflicting results for its functional activity but was previously described in an asymptomatic, homozygous child with a typical biochemical HPP signature (Saraff et al., 2016; Uday et al., 2019). The ALPL Gene Variant Consortium is currently performing in-depth investigation of this and other recurrent VUS for potential reclassification (Farman et al., 2024).
While our results improve the understanding of HPP molecular pathology, this study has some limitations. ALPL variant classifications are continuously being updated as new information emerges, as are the Global HPP Registry data. Data reported to the registry after the cutoff date for this analysis (September 2022) were not included. The nature of registry data and the process of resolving conflicting data entries inevitably lead to a lag between data cutoff dates and publication. An additional limitation is the lack of standardization in the method of gene sequencing for reported data, which reflects real-world practice; half the patients underwent single-gene sequencing, and the remainder underwent targeted, whole-exome, or panel sequencing or did not have a method specified. Not all sequencing methods have equivalent sensitivity to detect ALPL variants (Kishnani et al., 2017), and therefore some gene variants may have gone undetected. In addition, some haplotypes may have been interpreted as independent alleles, owing to the absence of parental segregation data. Our process for resolving conflicting variant entries may have introduced some unevenness in variant classifications, as variants with no conflicts between registry entries were not compared against the reference databases. We also note that, for the purposes of this analysis, perinatal/infantile HPP was defined as first manifestation of the disease occurring at age ≤6 months and we therefore cannot exclude the possibility that patients with prenatal benign HPP were included in this category. Additionally, the registry does not capture patient data posthumously. Therefore, the most severely affected patients may be underrepresented. It is also important to note that wide geographic areas, such as Africa and central and South America, are not currently represented in the Global HPP Registry. There are very few reports of HPP in Africa outside of South Africa and North Africa (Nunes, 2007). Others have noted ascertainment bias affecting interpretation of HPP prevalence throughout Africa. Lastly, some variants reported in the Registry may have been identified in screening of affected patients/families and may therefore overestimate the penetrance and/or severity of some genetic variants.
In conclusion, this analysis confirmed previously known ALPL variants and discovered novel variants in more than 800 patients from the Global HPP Registry. Variant numbers per patient, predicted functional consequences, and frequencies of variant types were predominantly similar across the geographic regions included in the Global HPP Registry, with the Japanese population containing both a higher proportion of patients with two or more variants and a disease onset before 6 months of age. A full understanding of the genetics of HPP across the geographic, ethnic, and age distribution of affected patients offers valuable insights into the pathogenesis and diagnosis of this rare disease.
AUTHOR CONTRIBUTIONS
All authors contributed to study design, served as study investigators, enrolled patients, collected and assembled data, provided data interpretation, and prepared, reviewed and revised, and approved the manuscript. SF performed data analysis.
ACKNOWLEDGMENTS
The authors gratefully acknowledge the patients and their families, clinical personnel, and all Global HPP Registry investigators, who contributed data on adults for these analyses, and the Global HPP Registry team.
FUNDING INFORMATION
This study was sponsored by Alexion, AstraZeneca Rare Disease (Boston, MA, USA). The Global HPP Registry is sponsored by Alexion, AstraZeneca Rare Disease and is overseen by a scientific advisory board comprising HPP clinical experts, including employees of Alexion, AstraZeneca Rare Disease. Alexion, AstraZeneca Rare Disease participated in the conception and design of the registry, collection of data, and the statistical analyses, as described in the Author Contributions section. Medical writing support in the development of this article was provided by Lela Creutz, PhD, and Drayton Hammond, PharmD, of Peloton Advantage, LLC, an OPEN Health company, and funded by Alexion, AstraZeneca Rare Disease.
CONFLICT OF INTEREST STATEMENT
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. PSK, LS, KMD, GAMM, AL, KO, WH, and CRG are consultants for and have received research funding and honoraria from Alexion, AstraZeneca Rare Disease. AP, WRM, and SF are employees of and may own stock/options in Alexion, AstraZeneca Rare Disease.
PATIENT CONSENT STATEMENT
All participants provided written informed consent prior to enrollment in the Global HPP Registry.
Open Research
DATA AVAILABILITY STATEMENT
Alexion, AstraZeneca Rare Disease will consider requests for disclosure of clinical study participant-level data provided that participant privacy is assured through methods like data de-identification, pseudonymization, or anonymization (as required by applicable law), and if such disclosure was included in the relevant study informed consent form or similar documentation. Qualified academic investigators may request participant-level clinical data and supporting documents (statistical analysis plan and protocol) pertaining to Alexion-sponsored studies. Further details regarding data availability and instructions for requesting information are available in the Alexion Clinical Trials Disclosure and Transparency Policy at https://alexion.com/our-research/research-and-development. Link to Data Request Form: https://alexion.com/contact-alexion/medical-information.




