RNA sequencing reveals deep intronic CEP120 variant: A report of the diagnostic odyssey for two siblings with Joubert syndrome type 31
There are many human disorders arising from the dysfunction of motile and/or primary cilia, collectively referred to as ciliopathies. The spectrum of phenotypes associated with ciliopathies includes intellectual disability and brain malformations, skeletal alterations, cystic kidneys, retinal degeneration, and infertility (Reiter & Leroux, 2017). Not only is there substantial clinical phenotypic overlap among ciliopathies, but variants within the same gene can also give rise to distinct ciliopathy syndromes. The CEP120 gene [MIM: 613446] is an example of this. More than 40 genes have been associated with Joubert syndrome (JBTS [MIM: PS213300]), including CEP120 (Joubert syndrome 31 (JBTS31) [MIM: 617761]), which is also associated with short-rib thoracic dysplasia 13 with or without polydactyly [MIM: 616300]. To date, 8 variants associated with JBTS31 have been reported in CEP120 within the literature, of which 7 are exonic; 5 missense, 1 nonsense, and 1 frameshift (Bowling et al., 2022; Roosing et al., 2016; Shaheen et al., 2015). The remaining variant is a 6 bp intronic deletion, 5 bp from the splice donor site of exon 2, which resulted in the use of an alternative splice site 483 bp downstream, predicted to result in a truncated protein. The use of this alternative splicing was seen in approximately 5% of mRNA transcripts, while the remaining mRNA had normal splicing from exon 2 to exon 3 (Roosing et al., 2016). Within ClinVar, one additional likely pathogenic variant in CEP120 has been reported to be associated with JBTS31 (as of October 13, 2023); this is a 2373 bp deletion (NM_001166226.1:c.-29-2113_128 + 103del), which includes all of exon 2 (Landrum et al., 2018). Here we present two brothers with a clinical diagnosis of Joubert syndrome, in whom we identified a novel missense variant and a deep intronic 57 bp deletion in CEP120 in trans. RNA sequencing (RNA-Seq) was key in the identification and interpretation of the deep intronic variant, leading to a clinical diagnosis.
The proband is an 18-years-old male with history of developmental delays, abnormal breathing patterns, abnormal eye movements, molar tooth sign on MRI, and multiple congenital anomalies, resulting in a clinical diagnosis of classic Joubert syndrome (Parisi, 2019). He is the first child of healthy non-consanguineous parents and was born at term with normal growth parameters after an uneventful pregnancy. The proband had a midline cleft palate that was repaired at 1 year of age. Abnormal breathing patterns with alternative periods of tachypnea with short periods of apnea were observed occasionally. He had feeding issues as a child and was gastrostomy-fed until 7 years of age. He now has full oral feeding and has chronic constipation. At 16 years and 9 months old, his height was 159 cm (2%, Z = −2.01) and his weight was 49.6 kg (4%, Z = −1.79). He has bilateral retinal coloboma, nonaccommodative esotropia, and abnormal eye movements, described as jerk nystagmus with a rotary component. He had strabismus repair at 11 years of age. He has an unsteady ataxic gait and genu valgum. He needs lower leg braces to ambulate and uses a walker for long distances. He can speak in full sentences, but his speech is slurred and can be difficult to understand. He followed a regular academic curriculum and finished high school with some accommodation. A cerebral MRI performed at 1 year of age revealed a large fourth ventricle, deep interpeduncular fossa, hypoplasia of the cerebellar vermis, midline vermis cleft, and prominence of the superior cerebellar peduncles consistent with a molar tooth sign (Figure 1b). Echocardiogram revealed a secundum atrial septal defect, which closed spontaneously. His annual abdominal and renal ultrasounds, as well as renal and liver function tests, are normal.
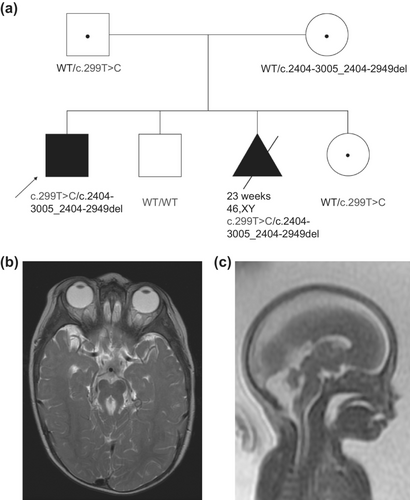
The proband's parents had three subsequent pregnancies, one affected and two unaffected (Figure 1a). Molecular prenatal diagnostic options were not available as the underlying etiology for the proband's Joubert syndrome had not been yet identified; surveillance with fetal ultrasounds and MRI were pursued. Fetal MRI at 22 weeks of gestation for the couple's third pregnancy revealed a male with bilateral club feet, cleft lip as well as cerebellar vermis hypoplasia, squaring of the fourth ventricle, elongation of the superior cerebellar peduncles and elongation of the brain stem, findings highly suggestive of Joubert syndrome (Figure 1c), and the pregnancy was interrupted shortly thereafter.
The proband's clinical testing included single gene testing for Joubert syndrome (CEP290, NPHP1, AHI1, MKS3, and TMEM67 genes) in 2006–2007 and a Joubert syndrome panel in 2012, all of which were non-diagnostic. Chromosomal microarray was normal. The family was subsequently enrolled in the Care4Rare Canada research program in 2014. Study design approval was obtained from the institutional research ethics board (Children's Hospital of Eastern Ontario; CTO-1577) and the parents provided informed consent. Duo (proband and affected fetus) research exome sequencing (ES) analysis in 2014 identified GPRIN2 [MIM: 611240] compound heterozygous variants (NM_014696.3: c.10A > T p.(Ser4Cys) and c.329G > A p.(Arg110Gln)) in both affected as a weak novel candidate, as the gene product induces outgrowth of neurites (Chen et al., 1999). However, subsequent re-analysis of the ES data in 2019 no longer considered this a compelling gene candidate as the c.329G > A variant was seen in 30 individuals in our in-house database (∼2000 exomes previously sequenced at the McGill University and Genome Quebec Innovation Centre), has an allele frequency of 0.0037 in gnomAD, and in silico programs do not predict this to be damaging (SIFT 0.32, PolyPhen 0.027, and no CADD score).
From this 2019 ES re-analysis, a gene candidate of SLC9A7 [MIM: 300368] was proposed. Both affected siblings were hemizygous for a maternally inherited (NM_001257291.1) c.923C > T p.(Ala308Val) variant which segregated appropriately in the unaffected male sibling, was not seen hemizygously in gnomAD and was absent from our in-house database. A report was published in 2018 demonstrating a recurrent de novo SLC9A7 missense variant (c.1543C > T p.(Leu515Phe)) was associated with multigenerational nonsyndromic intellectual disability in two unrelated families, with clinical presentations including early hypotonia and gross motor delay [MIM: 301024] (Khayat et al., 2018). Phenotypically, this was deemed not a good fit with our affected brothers, who have classic Joubert syndrome-like features. In this same analysis, a CEP120 missense variant (c.299 T > C; p.(Ile100Thr) (NM_001166226.1)) carried by both affected individuals was also proposed as a candidate. The variant was not seen previously in our in-house database, is absent from gnomAD and ClinVar, and is predicted deleterious by in silico programs (SIFT 0, PolyPhen 0.486, CADD 27.8). At the time of the analysis, CEP120 was deemed a very good phenotypic fit as it is associated with JBTS31; however, only one variant was identified by ES and this is an autosomal recessive (AR) condition.
In 2020, quad-research genome sequencing (GS) was performed using samples from the proband, affected fetus, and parents. From this analysis, the missense variant in CEP120 was deemed the most interesting candidate and was seen to be paternally inherited. However, a maternally inherited variant of interest in CEP120 could not be identified; only 3 maternally inherited variants were called in the GS data, all of which were deep intronic and were lacking in both conservation and nearby regulatory elements.
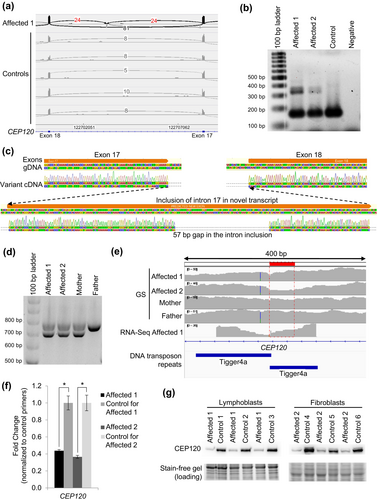
Research RNA-Seq was then performed using blood from the proband, collected in PAXgene Blood RNA tubes, to assess if there were any other candidate genes and investigate the possibility of a second causative variant in CEP120. From these analyses, there were instances of inclusion of intron 17 of CEP120 in some transcripts in the affected individual (Figure 2a). One of these junctions (chr5:122703395-122708343; GRCh37 reverse strand) was flagged in 1 of our 263 controls (z = 3.16), while the other junction of this intron inclusion (chr5:122700289-122703175) was not (z = inf); this splice event has not been identified in any other Care4Rare samples sent for RNA-Seq (n = 101 for blood). This aberrant transcript was confirmed in lymphoblast cells derived from the proband as well as in fibroblast cells derived from the affected fetus (Figure 2b). Sanger sequencing of the larger cDNA product revealed a 57 bp deletion of the DNA sequence within the novel intron inclusion, which was not anticipated (Figure 2c). Investigation of the GS data at and surrounding this location yielded no information regarding the DNA variant leading to this intron inclusion. We then performed PCR of the genomic DNA (gDNA) surrounding the intron inclusion, and from this saw that both affected and their mother carry a deletion on one allele (Figure 2d). Sanger sequencing of the gDNA identified a maternally inherited c.2404-3005_2404-2949del in both affected siblings, which correlates with the 57 bp deletion originally identified by Sanger sequencing the cDNA, and has an allele frequency of 0.000415 in gnomAD and is absent from ClinVar. Segregation of the paternally inherited c.299 T > C allele and this maternally inherited c.2404-3005_2404-2949del allele confirmed that these variants were only both present in the affected, while unaffected siblings carried either one or neither of these variants (Figure 1a). This deletion was not called by both our GS and RNA-Seq pipelines because it is in a highly repetitive region where there are tandem Tigger4a repeats, resulting in read misalignment. However, some dips in coverage can be seen in both GS and RNA-Seq datasets within this region (Figure 2e). To investigate the functional impacts of the c.2404-3005_2404-2949del; p.(Lys801_Val802ins*42) allele, RT-qPCR to assess transcript levels and western blot analyses of whole lymphoblast and whole fibroblast cell extracts was performed, which demonstrated markedly decreased CEP120 transcript and protein expression in the cells from both affected siblings (Figure 2f, g) (Lin et al., 2013), suggesting that there is mRNA nonsense-mediated decay with subsequent loss of protein expression, in line with a loss-of-function disease mechanism.
Overall, our patients' clinical presentations have significant overlap with other reported cases of JBTS31. After a more than decade-long diagnostic odyssey, including clinical ES, multiple re-analyses of ES data, GS, and RNA-Seq, we were finally able to identify the causative variants in this family. The compatibility with the autosomal recessive inheritance pattern, clinical findings, in silico predictions, RNA-Seq, and immunoblotting support classification of these c.299 T > C and c.2404-3005_2404-2949del CEP120 variants as likely pathogenic (PM2, PP3, PS3). This study highlights the benefits of RNA-Seq in obtaining a diagnosis for patients with rare diseases where additional data is required after inconclusive clinical testing, particularly in instances like this where variants have an impact on mRNA splicing and may have been undetected or missed by ES and/or GS. Therefore, the adaptation of RNA-Seq as a second-tier clinical test should be considered when there is a strong suspicion of a causative gene but a lack of a molecular diagnosis.
AUTHOR CONTRIBUTIONS
Aren E. Marshall and Gabrielle Lemire drafted the article and Kym M. Boycott and Kristin D. Kernohan revised the article for intellectual content. Aren E. Marshall and Jorge Davila had major roles in acquisition of the data. Aren E. Marshall, Yijing Liang, and Madeline Couse analyzed the data. Aren E. Marshall, Kym M. Boycott and Kristin D. Kernohan interpreted the data.
ACKNOWLEDGMENTS
We thank the family for their participation. We thank Tang K. Tang, PhD (Institute of Biomedical Sciences, Academia Sinica) for providing the CEP120 antibody. This study was performed under the Care4Rare Canada Consortium funded by Genome Canada and the Ontario Genomics Institute (OGI-147), the Canadian Institutes of Health Research, Ontario Research Fund, Genome Alberta, Genome British Columbia, Genome Quebec, and Children's Hospital of Eastern Ontario Foundation. We would like to acknowledge Wendy Mears for her technical assistance. A.E.M. was supported by a Canadian Institutes of Health Research (CIHR) fellowship award (MFE-176616). K.M.B. was supported by a CIHR Foundation Grant (FDN-154279) and a Tier 1 Canada Research Chair in Rare Disease Precision Health.
FUNDING INFORMATION
Canadian Institutes of Health Research, Grants/Award Number: FDN-154279; Children's Hospital Academic Medical Organization through Children's Hospital of Eastern Ontario; Ontario Genomics Institute, Grant/Award Number: OGI-147.
CONFLICT OF INTEREST STATEMENT
The authors have no conflict of interest to declare.
Open Research
DATA AVAILABILITY STATEMENT
Care4Rare Canada deposits multi-omic data in a national platform, Genomics4RD (genomics4rd.ca), to facilitate data sharing. Phenotypic and DNA/RNA sequencing data from this family is made available through a controlled access request to Genomics4RD ([email protected]).




