Time to diagnosis in rapid exome/genome sequencing in the clinical inpatient setting
Abstract
Exome and genome sequencing are clinically available, with many laboratories offering expedited testing (e.g., “rapid” and “ultra-rapid”). With the increase in uptake of expedited testing, there is a need for the development of inpatient protocols for best practices based on real-life data. A retrospective 2-year review (October 2019–November 2021) of the utilization of rapid exome and genome sequencing for inpatient cases at a tertiary care center using a utilization management tracking database with subsequent chart review was performed. Thirty-three expedited “rapid/priority” exome/genome tests were performed clinically. The average total turnaround time (TAT) was 17.88 days (5–43 days) with an average TAT of 13.97 days (3–41 days) for the performing laboratory. There were 5 positive diagnostic results (15.2%), 3 likely positive diagnostic results (9%), 2 noncontributory results (6%), and 26 nondiagnostic results (69.7%). Real-life data suggest that there is an approximately 3.91-day lag in getting samples to the performing laboratory. Although laboratories may advertise their expected TAT, a number of factors can potentially impact the actual time from test order placement to communication of the results for clinical use. Understanding the points of delay will enable the development of internal protocols and policies to improve time to diagnosis.
1 INTRODUCTION
The outcomes and potential benefits of rapid exome and genome sequencing have been analyzed in many research settings (Stark & Ellard, 2022; Vears et al., 2023). Among the benefits, specifically in pediatric critical care populations, are improvements in clinical outcomes with genetic results leading to change in management including transition to palliative care, a shift to precision treatments, early identification of comorbidities associated with diagnoses, and avoidance of invasive procedures (Australian Genomics Health Alliance Care Flagship, et al., 2020; Beaman et al., 2022; D. P. Dimmock et al., 2020; D. Dimmock et al., 2021; Elliott et al., 2019; Farnaes et al., 2018; Freed et al., 2020; Kingsmore & Cole, 2022; Lumaka et al., 2023; Meng et al., 2017; NICUSeq Study Group et al., 2021; Sanford et al., 2019; Stark et al., 2018; Sweeney et al., 2021; Wu et al., 2021). In addition, despite the relatively high cost of genetic sequencing compared to many other diagnostic tests, the use of broad genetic testing such as exome and genome sequencing has been reported to reduce healthcare-associated costs (Diaby et al., 2022; D. Dimmock et al., 2021; Farnaes et al., 2018; Incerti et al., 2022; Kingsmore & Cole, 2022; E. S. Kobayashi et al., 2022; E. F. S. Kobayashi & Dimmock, 2022; Stark et al., 2018; Stark & Ellard, 2022). One report illustrated increased cost savings as the turnaround time (TAT) decreased, with even a reduction of a few days making a significant difference (Kingsmore & Cole, 2022). When surveyed, these savings are typically concentrated in the areas of hospital admission costs and the cost of invasive interventions (Diaby et al., 2022; D. Dimmock et al., 2021; Farnaes et al., 2018; Kingsmore & Cole, 2022; E. F. S. Kobayashi & Dimmock, 2022; Stark et al., 2018).
Because both patient outcomes and healthcare cost-associated savings are predicted to improve as time for lab-reported results decreases for inpatients (Kingsmore & Cole, 2022), accelerating the TAT of genetic testing for this patient population becomes a priority. The TAT specific to each expedited sequencing test is typically stated by the individual laboratory (Kingsmore et al., 2019); however, the time associated with other factors for the decision to test and testing process has not been well described. Such factors could include genetic counseling, documentation of consent, sample collection from both patient and family member(s), and sample shipment. We aimed to examine the delays that may be associated with these steps in clinical practice, as well as identify other factors that may be contributing to TAT for expedited, more rapid, genetic testing.
2 METHODS
The Stanford University Medical Center includes Stanford Health Care and Stanford Children's Health, and the clinical laboratories serve both systems. Data were collected on expedited, more rapid, exome or genome sequencing ordered between October 2019 and November 2021. The tests were identified through the tracking database by Stanford Genetic Testing Optimization Service, a laboratory utilization management service. Based on quality improvement initiative, the institutional review board determined review was not required. Only inpatient orders were included.
Data included patient phenotype, type of test sent, care setting (e.g., inpatient, critical care), department/division of ordering provider, and outcome of testing. In addition, dates of interest collected included initiation of medical genetics consult (if applicable), testing recommendation, consent documentation, collection of patient sample, arrival of sample to testing lab, and return of result.
Using these data, we calculated the average number of days taken in each step in the clinical testing process. Day 0 was considered to be the day on which expedited testing was recommended. These calculations were performed for our data set as a whole as well as for the group of cases in which a positive/likely positive result was obtained.
3 RESULTS
A total of 33 expedited “rapid” exome or genome tests were performed during the period encompassed by this study. The tests were sent to one of seven commercial laboratories, hence representing a broad range of laboratories. Advertised average TAT collected from company websites as of January 2023 for the most rapid expedited test for all laboratories ranged from 3 to 14 days; the advertised TAT at the time of testing is unknown. Twenty-eight tests were ordered either by or in consult with the Stanford Medical Genetics team, and the remaining five tests were sent by Cardiology, Allergy and Immunology, Hematology and Oncology, Pediatric Intensive Care, and Cardiovascular Intensive Care teams.
There was a positive or likely positive diagnostic yield of 24.2%. There were 5 positive diagnostic cases (15.2%), 3 likely diagnostic (i.e., likely pathogenic variant(s) that explained the patient's reason for testing) (9%), 2 noncontributory (e.g., rapid testing identified a pathogenic or likely pathogenic variant that was previously identified but did not explain the patient's reason for testing) (6%), and 26 nondiagnostic cases (69.7%). There were eight pathogenic or likely pathogenic variants thought to be diagnostic for the reason the test was ordered in the following genes with associated diagnoses noted: OTC (ornithine transcarbamylase deficiency; MIM: 311250), DNM1L (lethal encephalopathy due to defective mitochondrial peroxisomal fission; MIM: 614388), KCNJ11 (hyperinsulinemic hypoglycemia; MIM: 601820), MYH7 (hypertrophic cardiomyopathy; MIM: 192600), PRF1 (hemophagocytic lymphohistiocytosis; MIM: 603553), PHKA2 (glycogen storage disease, Type IX; MIM: 306000), EP300 (Rubinstein-Taybi, Type 2; MIM: 613684), and CHD7 (CHARGE syndrome; MIM: 214800). All positive diagnostic cases were ordered either by or in consult with the Medical Genetics clinical team.
The average total TAT for all cases was 17.88 days (range 5–43 days) with an average TAT of 13.97 days (range 3–41 days) for the performing laboratory (Figure 1). The average number of days to obtain and document consent was 0.67 (range 0–10 days), 1.45 days to collect the patient sample (range 0–11 days), and 1.79 days for the sample to arrive at the performing lab after being collected (range 1–5 days). There were 12 cases in which samples were sent over weekends and holidays; the average time samples took in transit to the lab was 2.25 days compared to the 1.79-day average for all samples. The inpatient provider-dependent portion of this process (i.e., test recommendation to patient sample collection) took an average of 2.12 days (range 0–11 days). This led to an average 3.91-day (range 1–12 days) total lag time from the recommendation of expedited genetic testing to the date the patient sample arrived in the testing lab.
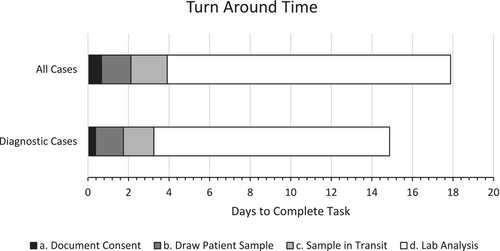
For the eight diagnostic/likely diagnostic cases in which a pathogenic or likely pathogenic variant was identified on expedited “rapid” testing, the average total TAT was 14.89 days (range 8–26 days) with an average lab analysis TAT of 11.63 days (range 7–22 days). In these cases, the average number of days to document consent was 0.38 days (range 0–3 days), with 1.38 days to collect the patient sample (range 0–3 days), and 1.5 days for the sample to arrive at the testing lab (range 1–5 days). For these cases, the average time from the recommendation of expedited “rapid” genetic testing to the date the patient sample was collected was 1.76 days (range 0–3 days).
4 DISCUSSION
The TATs of expedited “rapid” genetic testing reported by performing laboratories relate to the date of sample arrival to the date of result return (Kingsmore et al., 2019). The steps prior to the arrival of the patient sample can vary resulting in delays in diagnosis, which is particularly important in scenarios where more rapid testing is indicated. Delays in making a genetic diagnosis in this population may result in delays in appropriate care, more invasive testing strategies, longer hospital admissions, and increased costs associated with the hospitalization (Beaman et al., 2022; Diaby et al., 2022; D. Dimmock et al., 2021; Elliott et al., 2019; Farnaes et al., 2018; Freed et al., 2020; Incerti et al., 2022; Kingsmore & Cole, 2022; E. F. S. Kobayashi & Dimmock, 2022; Sanford et al., 2019; Stark & Ellard, 2022; Wu et al., 2021). The total amount of time elapsed between the decision to send expedited “rapid” genetic testing and the return of results has not been well documented in the literature.
This study showed an average TAT of 13.97 days for laboratory analysis. This time, as well as the time in which the sample is in transit to the laboratory, is typically outside of the ordering physician's control. TAT for laboratory analysis was variable even with the exact same test from the same laboratory; for example, the range of one expedited test from a single laboratory (n = 11) was 4–17 days. For these patients in the inpatient and critical care settings, we observed an average lag time of 2.12 days from the time expedited “rapid” testing was recommended to the time a patient sample was collected.
The exact reasons for delays in each of the steps before a laboratory received the sample are nuanced and difficult to determine, which is a limitation of the study. The delay in obtaining counseling and consent is likely multifaceted. One reason could be the difficulty of providers trying to coordinate visits with parents during a hospital admission. In some cases, parents may be overwhelmed in a hospital and intensive care setting resulting in delays before providing consent. As this study was conducted during the early use of expedited sequencing at this institution, some healthcare providers were likely unfamiliar with the use of these more rapid genetic testing options, which may have led to confusion and delay in both ordering the test and ensuring samples were correctly drawn and delivered. Other potential factors for delay include shipping schedules if tests were recommended on nights, weekends, and holidays. For example, in the 12 cases in which samples were sent over weekends and holidays, the average time samples took in transit to the lab was 2.25 days compared to the 1.79-day average for all samples. In addition, it is possible that reduced staffing and restrictions on family allowance at the bedside during the coronavirus disease-2019 (COVID-19) pandemic could have contributed to delays. Any delay in obtaining parental samples, which was not measured in this study, likely relates to many of the above issues in obtaining consent.
In examining the TAT for the eight cases in which a positive or likely positive diagnostic result was obtained, a slightly lower average time to complete each step in the process was observed. Reasons for this may include involvement of practitioners familiar with medical genetics early in the hospital course, suggesting the patient's phenotype was quickly suspected to be due to an underlying genetic etiology and genetic testing was prioritized. In addition, medical geneticists are more likely to be familiar with the counseling, consent, and ordering processes and provide detailed pertinent phenotypic information. The latter may also have contributed to a shorter laboratory testing time by helping with variant interpretation. This highlights the importance of including medical geneticists in the process of expedited exome/genome sequencing in the inpatient setting.
This study noted a positive diagnostic yield rate of 24.2%. This study consisted of a small number of cases (n = 33), but is relatively congruous with other case series (Beaman et al., 2022; Diaby et al., 2022; D. P. Dimmock et al., 2020; D. Dimmock et al., 2021; Farnaes et al., 2018; Freed et al., 2020; Kingsmore et al., 2019; Kingsmore & Cole, 2022; Meng et al., 2017; NICUSeq Study Group et al., 2021; Powis et al., 2020; Stark & Ellard, 2022; Wu et al., 2021). Five of the expedited genetic tests sent during our study period were not done in consult with a medical geneticist. If these five cases were excluded the diagnostic yield would have increased to 28.6%. Future monitoring of these data with additional cases will be helpful to see if this diagnostic yield is maintained or even increased as provider experience, best practices, and laboratory capabilities improve.
The use of expedited “rapid” exome and genome sequencing in the clinical settings is increasingly becoming more frequent, but further work is needed to establish criteria for optimal use of this rapid diagnostic technology. Identifying inefficiencies in the testing process from the clinical evaluation of the patient to the delivery of results to the clinical team will become increasingly helpful in order to design protocols to improve time to diagnosis and testing yield. Genomic technologies are powerful tools that will require effectively intervening at key steps in the process to minimize delays. Continued monitoring as part of quality improvement initiatives will likely help to identify those key steps in the process where interventions can be implemented to improve time to diagnosis for patients with genetic conditions.
AUTHOR CONTRIBUTIONS
Alison Schildt: Formal analysis; methodology; data curation; writing—original draft; review and editing. David Stevenson: Conceptualization; methodology; formal analysis; writing—review and editing. Linbo Yu: Conceptualization; methodology; data curation; project administration; formal analysis; writing—review and editing. Beatriz Anguiano: Data curation; methodology; project administration; formal analysis; writing—review and editing. Carlos Suarez: Conceptualization; methodology; formal analysis; writing—review and editing.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




