A medical odyssey of a 72-year-old man with Charcot–Marie–Tooth disease type 2 newly diagnosed with biallelic variants in SORD gene causing sorbitol dehydrogenase deficiency
Collaborators of the Undiagnosed Disease Network are present in Appendix.
Abstract
A 72-year-old man was referred to the Undiagnosed Diseases Network (UDN) because of gradual progressive weakness in both lower extremities for the past 45 years. He was initially diagnosed as having Charcot–Marie–Tooth disease type 2 (CMT2) without a defined molecular genetic cause. Exome sequencing (ES) failed to detect deleterious neuromuscular variants. Very recently, biallelic variants in sorbitol dehydrogenase (SORD) were discovered to be a novel cause of inherited neuropathies including CMT2 or distal hereditary motor neuropathy (dHMN) referred to as Sorbitol Dehydrogenase Deficiency with Peripheral Neuropathy (SORDD, OMIM 618912). The most common variant identified was c.757delG; p.A253Qfs*27. Through the Vanderbilt UDN clinical site, this patient was formally diagnosed with SORDD after the identification of homozygosity for the above SORD frameshift through UDN Genome Sequencing (GS). His medical odyssey was solved by GS and detection of extremely high levels of sorbitol. The diagnosis provided him the opportunity to receive potential treatment with an investigational drug in a clinical trial for SORDD. We suggest that similar studies be considered in other individuals thought to possibly have CMT2 or dHMN.
1 INTRODUCTION
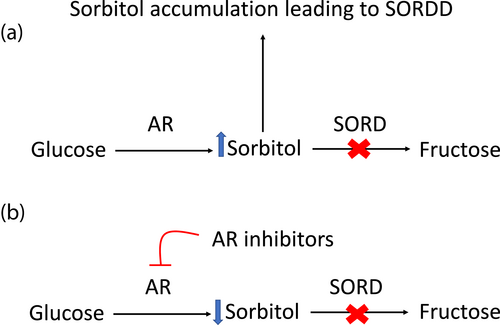
Aldose reductase converts glucose to sorbitol and Sorbitol dehydrogenase (SORD) converts sorbitol into fructose. Sorbitol dehydrogenase is a cytosolic enzyme encoded by the SORD gene. SORD deficiency with peripheral neuropathy (SORDD; OMIM 618912) is characterized by a progressive neuropathy that results in limb weakness and difficulty walking. The mean age of onset of neuropathy is late adolescence. In SORDD, the body is unable to metabolize sorbitol to fructose, which results in intracellular toxic sorbitol accumulation that causes symptoms of neuropathy. Previously, SORD deficiency was diagnosed symptomatically as Charcot–Marie–Tooth disease type 2 (CMT2) or distal Hereditary Motor Neuropathy (dHMN) because the association between neuropathy and SORD pathogenic variants was not known. However, a recent study discovered biallelic variants in SORD gene accounted for up to 10 percent of patients with CMT2 and dHMN. The variant c.757delG; p.A253Qfs*27 is reported to be the most common cause of SORDD and subsequently the most common cause of recessive neuropathy (Cortese et al., 2020). Currently, an investigational drug is being studied in patients with SORDD, which could potentially enable the development of future treatments for affected individuals suffering from neuropathy caused by SORDD.
2 CASE REPORT
The patient reported here is a 72-year-old male who presented with gradual progressive weakness in both lower extremities without sensory disturbance. He had a history of having weakness in both lower extremities since the age of 27. He has difficulty with ambulation leading to a slow gait and requires a cane for long walks. He has had numerous falls but without any serious injuries. His arm and hand strength are reduced, but he can perform his activities of daily living. He reported that he had several episodes of rhabdomyolysis after mountain climbing. He noted plantarflexor weakness at first greater than dorsiflexor weakness that was slowly progressive. Past medical history includes obesity, type 2 diabetes, and hypertension. Family history is unremarkable except for a history of tremulousness in a paternal aunt. Physical examination was notable for weakness in in both lower extremities without sensory loss. No pes cavus was noted on examination (Table 1). The first electromyography at the age of 27 was consistent with axonal neuropathy. A needle biopsy of lower extremity muscles at the age of 35 showed the absence of myopathy, which suggested a neurogenic disease. The primary diagnostic consideration at that time was CMT2. At the age of 54, he underwent spinal muscular atrophy testing, where SMN1 DNA sequencing was reported as normal. At the age of 65, the patient was screened using Invitae's CMT Disease Comprehensive panel that includes full-gene sequencing and deletion/duplication analysis of 57 genes (including PMP22) using next-generation sequencing technology, both which results were negative. At the age of 67, he underwent exome sequencing (ES), which was negative for deleterious neuromuscular variants. As such, he was referred to the Undiagnosed Diseases Network (UDN) and he was enrolled at the Vanderbilt UDN clinical site.
| OMIM reported features | Our patient | |
|---|---|---|
| Sex | Male 32/45 (71%) | Male |
| Age of onset (years) | 17 ± 8 (2–40) | 27 |
| Foot deformities | 31/45 (69%) | No |
| Distal lower limb weakness | 43/44 (98%) | Yes |
| Upper limb weakness | 26/44 (59%) | No |
| Foot plantar flexion involvement | 15/37 (41%) | Yes |
| Reduced vibratory sensation | 17/40 (42%) | No |
| Reduced pinprick superficial sensation | 13/39 (33%) | No |
| Tremor | 7/45 (15%) | No |
| Scoliosis | 4/45 (8%) | No |
| Hearing loss | 2/45 (4%) | No |
At 71 years of age when he was evaluated by the Vanderbilt UDN clinical site, it was noted that he had muscle weakness, atrophy, and steppage gait with numerous falls. Upper body strength was relatively unaffected compared to lower body. Sensation was intact. The remainder of his general physical examination was unremarkable. He underwent genome sequencing (GS). However on reanalysis a homozygous SORD frameshift variant (c.757delG:p.A253Qfs*27) was identified. His initial UDN GS report did not list his SORD variant. Although the variant was identified on this GS, there was no known disease association at the time of the reports. The variant was therefore not reported. However, on reanalysis once there was a known disease association therefore the variant was reported. Thus, he was formally diagnosed by the Vanderbilt UDN clinical site as having SORD due to homozygosity for a SORD frameshift variant. Thus the sorbitol level in his urine sample was then obtained and was found to be significantly elevated to greater than 1000 μM (normally hardly detectable in healthy individuals), which confirmed him as having SORDD.
3 DISCUSSION
Here we describe a patient with progressive neuropathy previously diagnosed as CMT2 without known molecular cause. He was found to have a newly identified homozygous variant in SORD that is associated with SORD with peripheral neuropathy (SORDD; OMIM 618912).
Charcot–Marie–Tooth (CMT) disease is characterized by hereditary sensory and motor neuropathies that damage the peripheral nerves. CMT type 1 (CMT1) is the most common type of CMT diseases and caused by damage to the myelin sheath while CMT2 is less common and caused by defects in the axon. The dHMN group of heterogenous inherited peripheral neuropathies overlaps both phenotypes and genotypes with CMT2 (Murphy et al., 2012). Genetic variants are found in more than 90% of the cases of CMT1, but genetic causes are undefined in up to 70% of CMT2 and dHMN cases (Fridman et al., 2014). However, very recently, biallelic variants in the SORD gene have been identified as the most common cause of undiagnosed cases of CMT2 and dHMN (Cortese et al., 2020). In our case, his progressive neuropathy, which had been symptomatically diagnosed as CMT2 with undefined genetic causes for the last 45 years, was finally diagnosed as SORDD through detection of a homozygous SORD pathogenic variant and elevated levels of sorbitol.
There are two ways to diagnose SORD deficiency. The first option is to measure sorbitol levels in a simple blood or urine sample. The second option is to perform genetic testing to detect biallelic pathogenic SORD variants. Variants in the SORD gene can cause a decrease in the levels and function loss of the SORD enzymes, leading to neuronal sorbitol accumulations (Stavrou et al., 2021). Cortese et al. demonstrated that fasting serum sorbitol concentrations were >100 times the upper limit of normal for patients who were homozygous for the p.A253Qfs*27 variant compared to the sorbitol concentrations in the control group (Cortese et al., 2020). In our patient's case, the sorbitol level in his urine sample was found to be significantly elevated to >1000 μM. His measurement of the sorbitol level was semiquantitative and not clinically reported, but the level was extraordinarily high. To our knowledge, there is no previous published correlation of sorbitol levels with disease expression. This test can be obtained easily at a low cost. Thus, we suggest that measurement of sorbitol levels in urine should be considered for patients with a symptomatic diagnosis of CMT2 or dHMN with an unknown genetic cause.
A recent study by Cortese et al reported that biallelic pathogenic variants in SORD with CMT2 or dHMN were identified, and that the majority of the disease causing variants were p.A253Qfs*27 (Cortese et al., 2020). Thus, it is important to identify this variant to be able to make a diagnosis of SORDD. However, the presence of a pseudogene makes it challenging to detect pathogenic SORD variants. The SORD gene has a highly homologous nonfunctional pseudogene, SORD2P. The c.757del variants are normally present in nonfunctional SORD2P pseudogene and are not disease causing. Thus, it is clinically important to differentiate the SORD gene from the SORD2P pseudogene (Laššuthová et al., 2021). Looking forward, next-generation sequencing will enable providers to identify this difference. Exome sequencing (ES) is an ideal molecular diagnostic method to detect such rare variants that can cause autosomal recessive and other Mendelian diseases (Elmas et al., 2019). In our case, the homozygous SORD frameshift variant was initially not detected by commercial ES. This was likely a consequence of the SORD2P pseudogene as has been suggested by the literature (Laššuthová et al., 2021). This variant was finally detected by UDN GS once a disease association was identified in the SORD gene on reanalysis. GS can detect copy number and noncoding variants as well as closely related sequences such as pseudogenes. Our case therefore demonstrates that the limited availability of GS in medical facilities likely results in missing variants outside exons as well as this variant likely due to the presence of a pseudogenes. In addition, our case also demonstrates the benefits of GS reanalysis after a period of time.
In patients with SORDD, the aldose reductase (AR) enzyme converts glucose to sorbitol, leading to a toxic accumulation of sorbitol (Figure 1). There are no approved drugs for SORD deficiency. However, patients who have been accurately diagnosed with SORDD have an opportunity to receive potential treatment with an investigational drug, govorestat that is currently being studied in a clinical trial. Applied Therapeutics, Inc has developed a central nervous system-penetrant AR inhibitor as known as AT-007 (govorestat), which is expected to treat SORDD (ClinicalTrials.gov Identifier: NCT05397665). Cortese et al. demonstrated that epalrestat and ranirestat, AR inhibitors, significantly reduced intracellular sorbitol accumulation in patient fibroblasts lacking functional SORD (Cortese et al., 2020). Additionally, long-term treatment with epalrestat was demonstrated to delay the progression of diabetic neuropathy and to have a favorable safety profile (Hotta et al., 2006). Sekiguchi et al. demonstrated ranirestat improved nerve conduction velocity in diabetic neuropathy (Sekiguchi et al., 2019). The current clinical trials of new drugs could provide an opportunity to receive potential treatment options for individuals with SORDD.
AUTHOR CONTRIBUTIONS
Yutaka Furuta drafted the article. John A. Phillips, III, Rizwan Hamid, Joy D. Cogan, Lynette Rives, and John H. Newman were responsible for the correct description display and interpretation of the genomic diagnostics and technical parts. Erica T. Nelson helped create figures. Serena M. Neumann, Erica T. Nelson, John A. Phillips, III, Rizwan Hamid, Rory J Tinker, and John H. Newman helped to draft the article. All authors read, corrected, and approved the final article.
ACKNOWLEDGMENTS
The authors are grateful to the patient for participating in the UDN. This work was supported in part by the NIH Common Fund, NIH/NHGRI grant 9U01NS134349-10 (JAP, JHN, and RH).
CONFLICT OF INTEREST STATEMENT
There are no conflicts of interest to declare.
APPENDIX A
Consortia: Collaborators of the Undiagnosed Disease Network (UDN) and the University of Miami include Maria T. Acosta, Margaret Adam, David R. Adams, Raquel L. Alvarez, Justin Alvey, Laura Amendola, Ashley Andrews, Euan A. Ashley, Carlos A. Bacino, Guney Bademci, Ashok Balasubramanyam, Dustin Baldridge, Jim Bale, Michael Bamshad, Deborah Barbouth, Pinar Bayrak-Toydemir, Anita Beck, Alan H. Beggs, Edward Behrens, Gill Bejerano, Hugo J. Bellen, Jimmy Bennett, Beverly Berg-Rood, Jonathan A. Bernstein, Gerard T. Berry, Anna, Bican, Stephanie Bivona, Elizabeth Blue, John Bohnsack, Devon Bonner, Lorenzo Botto, Brenna Boyd, Lauren C. Briere, Gabrielle Brown, Elizabeth A. Burke, Lindsay C. Burrage, Manish J. Butte, Peter Byers, William E. Byrd, John Carey, Olveen Carrasquillo, Thomas Cassini, Ta Chen Peter Chang, Sirisak Chanprasert, Hsiao-Tuan Chao, Ivan Chinn, Gary D. Clark, Terra R. Coakley, Laurel A. Cobban, Joy D. Cogan, Matthew Coggins, F. Sessions Cole, Heather A. Colley, Heidi Cope, Rosario Corona, William J. Craigen, Andrew B. Crouse, Michael Cunningham, Precilla D'Souza, Hongzheng Dai, Surendra Dasar, Joie Davis, Jyoti G. Dayal, Esteban C. Dell'Angelica, Patricia Dickson, Katrina Dipple, Daniel Doherty, Naghmeh Dorrani, Argenia L. Doss, Emilie D. Douine, Dawn Earl, David J. Eckstein, Lisa T. Emrick, Christine M. Eng, Marni Falk, Elizabeth L. Fieg, Paul G. Fisher, Brent L. Fogel, Irman Forghani, William A. Gahl, Ian Glass, Bernadette Gochuico, Page C. Goddard, Rena A. Godfrey, Katie Golden-Grant, Alana Grajewski, Don Hadley, Sihoun Hahn, Meghan C. Halley, Rizwan Hamid, Kelly Hassey, Nichole Hayes, Frances High, Anne Hing, Fuki M. Hisama, Ingrid A. Holm, Jason Hom, Martha Horike-Pyne, Alden Huang, Sarah Hutchison, Wendy Introne, Rosario Isasi, Kosuke Izumi, Fariha Jamal, Gail P. Jarvik, Jeffrey Jarvik, Suman Jayadev, Orpa Jean-Marie, Vaidehi Jobanputra, Lefkothea Karaviti, Shamika Ketkar, Dana Kiley, Gonench Kilich, Shilpa N. Kobren, Isaac S. Kohane, Jennefer N. Kohler, Susan Korrick, Mary Kozuira, Deborah Krakow, Donna M. Krasnewich, Elijah Kravets, Seema R. Lalani, Byron Lam, Christina Lam, Brendan C. Lanpher, Ian R. Lanza, Kimberly LeBlanc, Brendan H. Lee, Roy Levitt, Richard A. Lewis, Pengfei Liu, Xue Zhong Liu, Nicola Longo, Sandra K. Loo, Joseph Loscalzo, Richard L. Maas, Ellen F. Macnamara, Calum A. MacRae, Valerie V. Maduro, AudreyStephannie Maghiro, Rachel Mahoney, May Christine V. Malicdan, Laura A. Mamounas, Teri A. Manolio, Rong Mao, Kenneth Maravilla, Ronit Marom, Gabor MarthBeth A. Martin, Martin G. Martin, Julian A. Martínez-Agosto, Shruti Marwaha, Jacob McCauley, Allyn McConkie-Rosell, Alexa T. McCray, Elisabeth McGee, Heather Mefford, J. Lawrence Merritt, Matthew Might, Ghayda Mirzaa, Eva Morava, Paolo Moretti, John Mulvihill, Mariko Nakano-Okuno, Stanley F. Nelson, John H. Newman, Sarah K. Nicholas, Deborah Nickerson, Shirley Nieves-Rodriguez, Donna Novacic, Devin Oglesbee, James P. Orengo, Laura Pace, Stephen Pak, J. Carl Pallais, Christina G.S. Palmer, Jeanette C. Papp, Neil H. Parker, John A. Phillips III, Jennifer E. Posey, Lorraine Potocki, Barbara N. Pusey Swerdzewski, Aaron Quinlan, Deepak A. Rao, Anna Raper, Wendy Raskind, Genecee Renteria, Chloe M. Reuter, Lynette Rives, Amy K. Robertson, Lance H. Rodan, Jill A. Rosenfeld, Natalie Rosenwasser, Francis Rossignol, Maura Ruzhnikov, Ralph Sacco, Jacinda B. Sampson, Mario Saporta, Judy Schaechter, Timothy Schedl, Kelly Schoch, Daryl A. Scott, C. Ron Scott, Elaine Seto, Vandana Shashi, Jimann Shin, Edwin K. Silverman, Janet S. Sinsheimer, Kathy Sisco, Edward C. Smith, Kevin S. Smith, Lilianna Solnica-Krezel, Ben Solomon, Rebecca C. Spillmann, Joan M. Stoler, Kathleen Sullivan, Jennifer A. Sullivan, Angela Sun, Shirley Sutton, David A. Sweetser, Virginia Sybert, Holly K. Tabor, Queenie K.-G. Tan, Amelia L. M. Tan, Arjun Tarakad, Mustafa Tekin, Fred Telischi, Willa Thorson, Cynthia J. Tifft, Camilo Toro, Alyssa A. Tran, Rachel A. Ungar, Tiina K. Urv, Adeline Vanderver, Matt Velinder, Dave Viskochil, Tiphanie P. Vogel, Colleen E. Wahl, Melissa Walker, Stephanie Wallace, Nicole M. Walley, Jennifer Wambach, Jijun Wan, Lee-kai Wang, Michael F. Wangler, Patricia A. Ward, Daniel Wegner, Monika Weisz Hubshman, Mark Wener, Tara Wenger, Monte Westerfield, Matthew T. Wheeler, Jordan Whitlock, Lynne A. Wolfe, Kim Worley, Changrui Xiao, Shinya Yamamoto, John Yang, Zhe Zhang, Stephan Zuchner.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




