Tissue mosaicism, FMR1 expression and intellectual functioning in males with fragile X syndrome
Funding information: Chile's National Commission for Scientific and Technological Research; Comision Nacional de Investigacion Cientifica Y Tecnologica; Financial Markets Foundation for Children, Grant/Award Number: 2017-361; International Postgraduate Research Scholarship; Martin & E.H. Flack Trust; Medical Research Future Fund, Grant/Award Number: 1141334; Murdoch Children's Research Institute; National Health and Medical Research Council, Grant/Award Numbers: 1103389, 1049299; Pierce Armstrong Trust; Research Training Program Scholarship; Royal Children's Hospital Foundation; The Victorian Government's Operational Infrastructure Support Program; NHMRC Early Career Fellowship, Grant/Award Number: 1112934
Abstract
Fragile X syndrome (FXS) is caused by hypermethylation of the FMR1 promoter due to the full mutation expansion (full mutation [FM]: CGG ≥ 200 repeats) and silencing of FMR1. Assessment of mosaicism for active-unmethylated alleles has prognostic utility. This study examined relationships between FMR1 methylation in different tissues with FMR1 messenger ribonucleic acid (mRNA) and intellectual functioning in 87 males with FXS (1.89–43.17 years of age). Methylation sensitive Southern blot (mSB) and Methylation Specific-Quantitative Melt Aanalysis (MS-QMA) were used to examine FMR1 methylation. FMR1 mRNA levels in blood showed strong relationships with FMR1 methylation assessed using MS-QMA in blood (n = 68; R2 = 0.597; p = 1.4 × 10−10) and buccal epithelial cells (BEC) (n = 62; R2 = 0.24; p = 0.003), with these measures also showing relationships with intellectual functioning scores (p < 0.01). However, these relationships were not as strong for mSB, with ~40% of males with only FM alleles that were 100% methylated and non-mosaic by mSB, showing methylation mosaicism by MS-QMA. This was confirmed through presence of detectable levels of FMR1 mRNA in blood. In summary, FMR1 methylation levels in blood and BEC examined by MS-QMA were significantly associated with FMR1 mRNA levels and intellectual functioning in males with FXS. These relationships were not as strong for mSB, which underestimated prevalence of mosaicism.
1 INTRODUCTION
Fragile X syndrome (FXS) is a common form of inherited intellectual disability (ID), reported to affect 1 in 3600 males and 1 in 6000 females in the general population (Hagerman et al., 2009). It is caused by a trinucleotide expansion, ≥200 CGG repeats in the 5′ untranslated region of the Fragile X Messenger Ribonucleoprotein 1 (FMR1) gene, called full mutation (FM) (Verkerk et al., 1991). FM alleles are usually associated with abnormal DNA methylation of the FMR1 promoter, with consequent silencing of FMR1 messenger ribonucleic acid (mRNA) and its protein (FMRP; reviewed in Kraan et al. [2019]). The absence of FMRP is associated with the characteristic features of FXS, including learning and memory deficits, ID, behavioral problems, and autism features (Hagerman & Harris, 2008).
The FMR1 gene shows a high degree of somatic instability, such that many individuals have mosaic alleles of different CGG size and/or methylation within or between different tissues (Stoger et al., 2011; Tassone et al., 2000). For instance, some cells may have methylated and inactive FM alleles, while others may carry unmethylated and active alleles of different sizes, including normal (<44 CGG), intermediate (44–54 CGG), and premutation size (PM: 55-199CGG), with their proportions often varying between tissues. Previous studies have also reported variability in DNA methylation at different regions of the FMR1 promoter, between CpG sites, between sexes and over time (Aliaga et al., 2016; Godler et al., 2013; Stoger et al., 2011). Despite this complexity, these individuals are commonly grouped in the literature under the same “umbrella” of FMR1 methylation or CGG size mosaicism, with contribution of tissue mosaicism to FXS severity not well understood (Nolin et al., 1994; Pretto et al., 2014; Rousseau et al., 1991a).
Technical limitations of methods used to detect mosaicism is another issue. The methylation and/or CGG size mosaicism for PM and FM alleles has been reported to vary between 12% (Rousseau et al., 1991a) and 41% (Nolin et al., 1994) in males with FM identified using the standard two-step protocol (Biancalana et al., 2015). However, these figures may be under-estimates, as they are based on methylation sensitive Southern blot (mSB) that can fail to detect methylation or CGG size mosaicism in individuals who carry large FMR1 expansions in a small proportion of cells (<20%) (Aliaga et al., 2016). To address this problem, “long range” polymerase chain reaction (PCR) has been introduced. This PCR based approach has a higher analytical sensitivity than mSB and can amplify alleles up to FM size, directly targeting the CGG expansion. However, this also makes the approach vulnerable to: (i) preferential amplification of smaller alleles in individuals with mosaicism, decreasing analytical sensitivity for detection of mosaic, larger size CGG expansions (Aliaga et al., 2016; Field et al., 2019; Hensel et al., 2019); (ii) loss of PCR primer sites flanking CGG expansion (with some alleles not detected by PCR), reported to occur as part of somatic instability of these alleles in some individuals with CGG size mosaicism (Hwang et al., 2016).
To mitigate these issues, we have developed Methylation Specific-Quantitative Melt Analysis (MS-QMA) for detection of methylated FMR1 alleles. MS-QMA targets DNA methylation at the FMR1 exon 1/intron 1 boundary (downstream of the CGG repeat) and is therefore not subject to CGG size amplification bias, associated with CGG based screening, nor to loss of primer binding sites proximal to the CGG expansion as part of somatic instability. We used MS-QMA as a first-tier test on 3340 males with developmental delay referred for FXS testing (Aliaga et al., 2016), which increased FXS diagnostic yield by 15% (Aliaga et al., 2016). In blood of females (Inaba et al., 2014) and males (Kraan et al., 2020) with FXS, MS-QMA analysis of FMR1 methylation showed strong relationships with intellectual functioning, suggesting prognostic utility. However, these studies did not examine differences in tissue mosaicism, or how these may impact FMR1 expression and phenotype severity as reflected by intellectual functioning.
This study aimed to address these gaps by examining relationships between the differences in levels of FMR1 promoter methylation between tissues, and how these relate to FMR1 expression and intellectual functioning in a large cohort of males with FXS. In this study the presence of mosaicism was defined based on FMR1 promoter methylation being less than 100% and/or FMR1 mRNA level being greater than zero. Low-level mosaicism was defined as variability in methylation values not identified by mSB.
2 MATERIALS AND METHODS
2.1 Participants
The FXS cohort included 87 males from Australia (n = 43) and Chile (n = 44), 1.89–43.17 years of age. FM only (n = 70) and PM/FM (n = 17) allelic groups were defined based on allelic class provided to the family on the formal diagnostic report indicating presence or absence of PM together with FM alleles. The control cohort included 24 males aged 7–77 years with confirmed normal size alleles (<45 CGG).
For the Chilean and Australian cohorts, routine FXS testing involved first-tier PCR-based assessment of CGG repeat size with the upper limit of detection 330 CGG repeats for the Chilean cohort (Saluto et al., 2005) and 170 CGG repeats for the Australian cohort (Khaniani et al., 2008). Blood DNA samples with CGG size in the PM range or with failed PCR product, were referred for second-tier confirmatory testing by Southern blot. For the Australian participants Southern blot analysis did not utilize methylation sensitive restriction enzymes, as previously described (Francis et al., 2000). For the Chilean participants mSB was employed utilizing a different methodology (Alliende et al., 1998) to mSB in this study used for quantitative assessment of FMR1 methylation.
Australian FXS participants were recruited nationally through: (i) Victorian Clinical Genetics Services, Murdoch Children's Research Institute and Genetics of Learning Disability Service, Hunter Genetics; (ii) referring practitioners; and (iii) family support groups including the Fragile X Association of Australia. Chilean participants were recruited through Molecular and Cytogenetics at INTA, University of Chile. Individuals were excluded from the study if they had any other significant medical conditions (e.g., stroke, head trauma), any other genetic conditions of known clinical significance, and if they had inadequately controlled seizures. Table S1 provides age and intellectual functioning scores by allelic sub-group.
2.2 Procedure
Participants attended an appointment for the intellectual functioning assessment and collection of biological samples. For saliva collection, participants were required to spit directly into the tube to avoid contamination with buccal epithelial cells (BEC); however, not all participants were able to understand these instructions or could not spit, and thus no saliva sample was collected. The consent for the collection of venous blood was optional and not all parents/caregivers provided consent. In some cases where consent had been provided by the parent/caregiver, the child became distressed prior to the collection, or expressed dissent and thus collection was not performed. Similarly, some children became distressed during buccal collection and collection was ceased. Thus, not all participants provided all three tissue types for analysis and in some instances, there was not enough blood for FMR1 methylation and mRNA analyses. Table S2 provides a summary of available data (intellectual functioning and molecular variables) for each allelic group.
2.3 Ethics statement
This study was approved by the Royal Children's Hospital and INTA Research Ethics Committees (Single Site: HREC 34227A and HREC 33066F; Multi site HREC: HREC/13/RCHM/24; and INTA Human Research Ethics Committee #15). Signed informed consent was provided by the participants' legal representatives.
2.4 Assessment of intellectual functioning
Intellectual functioning of participants aged 3 years and older was assessed with one of the following standardized assessments depending on their age and country of residence: the Wechsler Preschool and Primary Scale of Intelligence—3rd Edition Australian and Mexican Editions (WPPSI-III; children aged 3–6 years) (Wechsler, 2002, 2004), the Wechsler Intelligence Scale for Children—4th Edition Australian (WISC-IV; Australian children aged 7–16 years) (Wechsler, 2003)/Wechsler Intelligence Scale for Children—3rd Edition Chilean version (WISC-III; Chilean children aged 7–16 years) (Wechsler, 2007) or the Wechsler Adult Intelligence Scale—4th Edition Australian and Chilean Editions (WAIS-IV; ≥17 years) (Wechsler, 2008a, 2008b). In this study we used both standard intelligence quotient (IQ) scores and corrected IQ scores (see Arpone et al., 2018 for a summary). The corrected IQ scores method reduces loss of data due to invalid scores and has been shown to result in a normal distribution of scores (Arpone et al., 2018).
2.5 Sample processing
All biological samples were collected at the time of assessment. Eight milliliters (ml) of blood was collected in ethylenediamine tetraacetic acid (EDTA) tubes: (i) 3 ml used for DNA extractions; (ii) 5 ml used for peripheral blood mononuclear cell (PBMC) isolation using Ficoll gradient separation, as previously described (Loesch et al., 2011). Approximately one million of isolated PBMCs per participant, were lysed in RLP-2-Mercaptoethanol buffer (Qiagen, Hilden, Germany), prior to total RNA being extracted using the RNeasy kit (Qiagen, Hilden, Germany) and stored at −80°C as per manufacturer's instructions (Qiagen, Hilden, Germany). Saliva and BEC samples were collected using the Oragene® DNA Self-Collection Kit (DNA Genotek, Global) and Master Amp Buccal Swab Brush kit (Epicenter technologies, Global), respectively. Up to four BEC brush samples (Epicenter technologies, Global), and a single saliva swab (DNA Genotek, Global) were collected per participant, and processed as per manufacturer's instructions. Brush and swab samples were inspected for blood contamination by at least two staff members at the time of sample collection, and/or at the time of sample receipt prior to processing. Collected brushes and swabs with confirmed blood contamination were discarded.
DNA was then extracted from blood, BEC, and saliva samples using NucleoSpin®Tissue genomic DNA extraction kit, as per manufacturer's instructions (MACHEREY-NAGELGmbH & Co. KG, Düren, Germany). Total RNA extraction was performed on PBMC lysates using the RNA easy Minikit (Qiagen, Hilden, Germany) according to the manufacturer's specifications. RNA quality and concentration were assessed by NanoDrop 2000 spectrophotometer.
2.6 Methylation Specific-Quantitative Melt Analysis (MS-QMA)
About 100–150 ng of DNA for each sample was transferred to 96-well plates to be treated with sodium bisulphite, as previously described (Inaba et al., 2014). Each DNA sample was bisulphite converted using the EZ DNA Methylation-GoldTM kit (Zymo research, Global) in two separate reactions, with each conversion analyzed in duplicate reactions using MS-QMA. Total 96 samples were bisulfite converted at a time (3 controls and 93 unknown samples per plate) and were serially diluted two times post-conversion. Each set of four 96 well plates was then transferred into a 384 well format for real-time PCR analysis utilizing MeltDoctor™ high-resolution melt reagents in 10 μl reactions, as per manufacturer's instructions (Life technologies, Foster City, CA). A unique primer set was used that targets methylation of specific CpG sites within the fragile X-related epigenetic element 2 (FREE2) region that were (Appendix 1) previously shown to be significantly associated with intellectual functioning in FXS (Godler et al., 2012). The annealing temperature for the thermal cycling protocol was 65°C for 40 cycles. The ViiA™ 7 Real-Time PCR System (Life technologies, Foster City, CA) was then used to measure the rate of dye incorporation into double stranded DNA in order to quantify DNA concentration of the unknown samples using the relative standard curve method. The dynamic linear range (between 0.05 and 10 ng/μl) was determined from the standard curve using a series of doubling dilutions of a converted DNA standard from a control lymphoblast cell line during each run. To progress to the next stage of the analysis the unknown samples had to be within this dynamic linear range.
The high-resolution melt step followed the real-time PCR in close tube format. The products from methylated and unmethylated FREE2 sequence were then separated into single strands in the temperature range of 74 and 82°C. The HRM Software Module for ViiA™ 7 System was then used to plot the rate of PCR product separation to single strands at different temperatures with the difference in fluorescence converted to Aligned Fluorescence Units (AFU) at 78°C. The AFU conversion to the methylation percentage, and all of the above quality control steps, were analyzed simultaneously for 384 reactions at a time using Q-MAX software (Curve Tomorrow, Melbourne, Australia), developed to automate the process (Inaba et al., 2014).
2.7 Methylation sensitive Southern blot analysis
Methylation sensitive Southern blot analysis of the FMR1 CpG island was performed as previously described (Hwang et al., 2016; Rousseau et al., 1991b). Specifically, 7–9 μg of DNA extracted from blood were double digested with NruI and HindIII methylation sensitive restriction enzymes. Blots were probed with P32 and labeled with pfxa3 using the Random Primer DNA labelling kit (Takara; Shiga Prefecture, Japan). NruI digestion methylation ratios (MRs) were quantified by densitometry scanning of bands produced by Typhoon Trio using Image Quant TL-software 7.0, with methylated and unmethylated FMR1 alleles discriminated based on their size, as previously described (Hwang et al., 2016). MRs were calculated as the total portion of the signal of the methylated band multiplied by 100 and divided by total signal from unmethylated and methylated bands with raw data from densitometry presented in Supporting Information in Appendix S1.
2.8 FMR1 mRNA analysis
Around 10 ng of RNA were used for complementary DNA (cDNA) strand synthesis using the High Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific, Global). FMR1 mRNA analysis was performed using the reverse transcription real-time PCR on a ViiA 7 Real-Time PCR System (Life Technologies, Global). The relative standard curve method was utilized for FMR1 5′ and 3′ mRNA quantification, with average of the two assays normalized to mRNA of two internal control genes (EIF4A2 and SDHA), as previously described (Kraan et al., 2016, 2018).
2.9 Statistical analysis
Summary statistics for age and standard and corrected IQ scores were presented by mean and standard deviation. Comparison of the difference in age between the groups (FM only and PM/FM mosaic) was conducted using two-sample t-test, while robust regression was used for corrected and standard IQ scores, with allelic group as a binary variable and adjusted for age and country whenever significant. Spearman's rank correlation was used to establish the relationship among DNA methylation variables, assessed using the MS-QMA and mSB methods, in whole sample and separately for FM only and PM/FM allelic groups, and in a sub-sample of individuals who had all molecular data available. These molecular variables were then used as the predictors for the relationships with FMR1 mRNA levels, as raw and log transformed data. Because the relationship may be different above or below a certain value of DNA methylation we used the piecewise linear regression to determine the breakpoint and the relationship below and above breakpoint. The analyses were conducted for all FXS data, and separately for the FM only and PM/FM allelic groups, and in the sub-sample where individuals have all molecular data. If no breakpoint was found, the robust regression method was used to assess the relationship. This method was also used for the relationship between standard IQ scores and all molecular variables, adjusted for country. However, because corrected IQ scores were non-linearly related to age, the semi-parametric regression was employed, with age modeled as non-parametric component and country and each molecular variable as parametric component. False discovery rate (FDR) was used for adjusting multiple testing. Analyses with log transformed FMR1 mRNA are also presented. Log transformation removes zero values and allows for visual inspection of variability in FMR1 expression.
All analyses were carried out using Stata package, version 15.0 (http://www.stata.com) and p-value (p) < 0.05 considered significant.
3 RESULTS
3.1 Intergroup comparisons for FMR1 mRNA and methylation measures
FMR1 mRNA levels were greater than zero in 57.9% of participants in the FM-only (33 out of 57 males) and 100% of PM/FM (n = 13) allelic groups providing evidence for presence of mosaicism (Figure 1a). For the combined cohort of 70 males with FXS (FM-only and PM/FM allelic groups), 68.6% (48 males) had FMR1 mRNA levels greater than zero in blood.
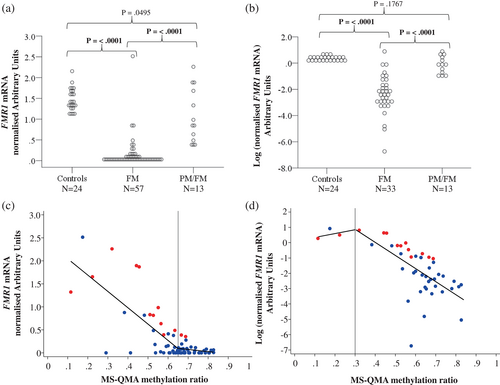
The FM only group had significantly lower FMR1 mRNA levels compared to the PM/FM allelic group, and male controls (Figure 1a). In contrast, the PM/FM group did not differ significantly from controls on FMR1 mRNA levels. To improve the visualization of the variability of FMR1 mRNA data with low expression values in the FM-only group, log transformation of the FMR1 mRNA data was performed and plotted (Figure 1b). This showed significant variation in FMR1 mRNA levels in the FM-only group, not obvious from the non-transformed data in Figure 1a. Although the FMR1 mRNA levels were significantly decreased in these individuals with FM as compared to controls, they were not significantly different in the FM only group between males with mSB methylation being less than or equal to 100% (Appendix 2), suggesting that mSB did not capture this biological variability in FMR1 expression in blood in the FM only group.
DNA methylation of <100%, was detected by mSB analysis in blood for 70.2% (40 out of 57) of males with FXS from the combined cohort (that had sufficient DNA available for mSB). When separated based on allelic groups, 62.2% (28 out of 45) of FM-only males and all (n = 12) PM/FM allelic group males had methylation less than 100% by mSB. In contrast to mSB analysis, all individuals with FXS showed MR less than 1 (equivalent to 100% by mSB) by MS-QMA in blood, BEC, and saliva DNA (Figure 1c,d).
3.2 Relationships between FMR1 methylation and mRNA levels
To confirm biological significance of the variability in methylation detected by MS-QMA we examined relationships between MS-QMA MR and FMR1 mRNA levels. Significant relationships were identified (Figure 1c,d), which were linear but with breakpoints present for blood DNA at 0.30 and 0.65 MR for log- and non-transformed FMR1 mRNA values, respectively. While non-transformed FMR1 mRNA data showed significant relationships with MS-QMA MR in blood and BEC, log-transformed mRNA data (with all zero FMR1 mRNA values excluded) only showed a significant relationship with MS-QMA MR in blood (Table 1). Transformation of FMR1 mRNA values using natural log showed that low-level mosaicism identified by MS-QMA in blood, was strongly correlated with the low-levels of FMR1 mRNA in blood (Figure 2b; Table 1).
| Break point | Piecewise linear regression | |||||||
|---|---|---|---|---|---|---|---|---|
| Variable | n | Estimate | 95% CI | Threshold | Coeff. (β) | SE | p | R-square |
| Log transformeda | ||||||||
| Blood MS-QMAb | 45 | 0.30 | (−0.13, 0.73) | <0.30 | 2.38 | 16.6 | 0.887 | 0.455 |
| ≥0.30 | −8.51 | 1.75 | 2.0 × 10−5 | |||||
| Saliva MS-QMAb | 28 | 0.32 | (−0.29, 0.92) | <0.32 | 2.94 | 8.26 | 0.725 | 0.055 |
| ≥0.32 | −2.55 | 2.33 | 0.284 | |||||
| Buccal MS-QMA | 41 | 0.59 | (0.26, 0.92) | <0.59 | −1.50 | 2.36 | 0.528 | 0.154 |
| ≥0.59 | −6.10 | 4.75 | 0.207 | |||||
| mSB | 39 | NA | NA | NA | −0.003 | 0.011 | 0.815 | 0.002 |
| Non-transformed | ||||||||
| Blood MS-QMA | 68 | 0.65 | (0.56, 0.74) | <0.65 | −3.55 | 0.46 | 1.4 × 10−10 | 0.597 |
| ≥0.65 | −0.47 | 1.14 | 0.680 | |||||
| Saliva MS-QMA | 44 | 0.51 | (0.24, 0.78) | <0.51 | −1.67 | 0.89 | 0.067 | 0.114 |
| ≥0.51 | 0.58 | 1.40 | 0.681 | |||||
| Buccal MS-QMA | 62 | 0.63 | (0.38, 0.88) | <0.63 | −2.33 | 0.75 | 0.003 | 0.240 |
| ≥0.63 | −0.62 | 1.66 | 0.709 | |||||
| mSB | 54 | NA | NA | NA | −0.001 | 0.001 | 0.427 | 0.007 |
- Note: CI = confidence interval; Estimate = estimate threshold; n = sample size; NA = did not find breakpoint; p = p-value; SE = standard error. The values in bold indicate p-values (p) that remained 〈 0.05 after adjustment for multiple testing, using FDR.
- a Relationship with log transformed FMR1 mRNA values using linear or piecewise linear regression.
- b Threshold was not significant.
When stratified by allelic group the relationship between FMR1 methylation assessed using MS-QMA and FMR1 mRNA levels in blood was significant for both the FM-only and PM/FM allelic groups using both log- and non-transformed FMR1 mRNA levels (Tables S3 and S4). Additionally, MS-QMA in BEC was also significantly associated with FMR1 mRNA levels in the PM/FM allelic group. In contrast, mSB methylation outputs were not associated with FMR1 mRNA levels in the combined cohort or the FM-only allelic group. However, mSB methylation outputs were significantly associated with FMR1 mRNA levels in the PM/FM allelic group (Table S4) that showed much greater range and variability in mRNA levels than the FM-only group (Figure 1c,d). Analysis with a sub-sample of males with complete data on all molecular variables demonstrated similar results, as observed in the whole sample (Table 2), when using both log- and non-transformed FMR1 mRNA data (Table S5).
| mSB | Blood MS-QMA | Saliva MS-QMA | |||||||
|---|---|---|---|---|---|---|---|---|---|
| n | ρ | p | n | ρ | p | n | ρ | p | |
| Whole sample | |||||||||
| mSB | – | ||||||||
| Blood MS-QMA | 57 | 0.23 | 0.089 | ||||||
| Saliva MS-QMA | 37 | 0.06 | 0.722 | 44 | 0.25 | 0.100 | |||
| Buccal MS-QMA | 51 | 0.14 | 0.325 | 65 | 0.35 | 0.004 | 45 | 0.75 | <0.001 |
| FM only | |||||||||
| mSB | – | ||||||||
| Blood MS-QMA | 45 | 0.05 | 0.724 | ||||||
| Saliva MS-QMA | 30 | 0.05 | 0.812 | 36 | 0.19 | 0.269 | |||
| Buccal MS-QMA | 40 | 0.06 | 0.693 | 53 | 0.22 | 0.114 | 36 | 0.73 | <0.001 |
| PM/FM Mosaic | |||||||||
| mSB | – | ||||||||
| Blood MS-QMA | 12 | 0.94 | <0.001 | ||||||
| Saliva MS-QMA | – | – | – | – | – | – | |||
| Buccal MS-QMA | 11 | 0.56 | 0.071 | 12 | 0.73 | 0.007 | – | – | – |
- Note: n = sample size; ρ = Spearman's rank correlation; p = p-value. The values in bold indicate p-values (p) that remained 〈 0.05 after adjustment for multiple testing, using FDR.
- Analyses for saliva in the PM/FM mosaic group not included due to small sample size (n = 7).
- Abbreviation: FM, full mutation.
3.3 Assessment of tissue mosaicism using MS-QMA and mSB methylation analyses
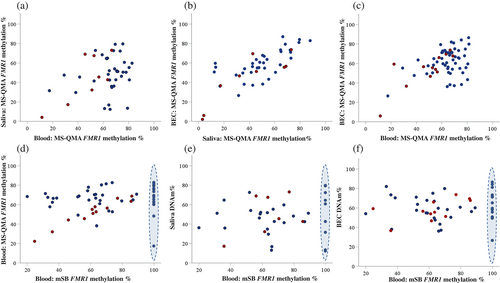
Percentage of FMR1 promoter methylation from MS-QMA (MR X 100) showed significant correlation between BEC with both saliva and blood. BEC and saliva FMR1 promoter methylation relationships were significantly stronger than the relationship between blood and BEC methylation (Figure 2; Table 2). Moreover, significant BEC versus saliva methylation relationships were present for both the FM-only and PM/FM allelic groups (Table 2). In contrast, significant relationships for BEC with blood DNA methylation were primarily driven by variability in the PM/FM allelic group and were absent for the FM-only group (Table 3). For mSB, only one significant relationship with MS-QMA methylation was evident in blood for the PM/FM allelic group, with no significant relationships observed between mSB in any tissue in the FM-only group (Table 2). A similar pattern was observed for the sub-sample of individuals who had a complete molecular data-set available for these analyses (Table S6). This lack of significant relationships for mSB may be explained by lack of variability in methylation values at the upper end of the distribution for a large proportion of samples in the FM-only group, as highlighted by blue ovals in Figure 2d–f. These samples showed variability in methylation % ranging from 20% to over 80%, when assessed using MS-QMA.
| Corrected IQ scores | ||||
|---|---|---|---|---|
| n | β | SE | p | |
| cFSIQ | ||||
| Blood MS-QMA | 69 | −47.91 | 13.99 | 0.001 |
| Saliva MS-QMA | 45 | −52.46 | 14.94 | 0.001 |
| Buccal MS-QMA | 73 | −37.95 | 13.25 | 0.006 |
| mSB | 53 | −0.109 | 0.101 | 0.287 |
| FMR1 mRNA | 66 | 10.46 | 3.715 | 0.006 |
| cVIQ | ||||
| Blood MS-QMA | 69 | −45.60 | 15.39 | 0.004 |
| Saliva MS-QMA | 45 | −52.05 | 16.60 | 0.003 |
| Buccal MS-QMA | 73 | −35.42 | 14.22 | 0.015 |
| mSB | 53 | −0.090 | 0.107 | 0.404 |
| FMR1 mRNA | 66 | 10.13 | 4.022 | 0.014 |
| cPIQ | ||||
| Blood MS-QMA | 71 | −44.99 | 14.33 | 0.002 |
| Saliva MS-QMA | 47 | −38.28 | 13.52 | 0.007 |
| Buccal MS-QMA | 75 | −29.96 | 13.10 | 0.025 |
| mSB | 55 | −0.124 | 0.095 | 0.198 |
| FMR1 mRNA | 68 | 9.528 | 3.765 | 0.014 |
- Note: n = sample size; β = estimated regression coefficient; p = p-value; SE = standard error. p-value (p) in bold remained < 0.05 after adjustment for multiple testing, using false discovery rate.
- Semi-parametric regression was used for corrected IQ scores, adjusted for age (non-parametric) and country (parametric).
3.4 Epi-genotype–phenotype studies
The FM-only group had significantly lower corrected Verbal IQ (cVIQ), Performance IQ (cPIQ) and Full Scale IQ (cFSIQ), and standard FSIQ scores compared to the group mosaic for PM and FM alleles. The two groups did not significantly differ on age (Table S1).
Analysis of FMR1 promoter methylation using MS-QMA in all three tissues and FMR1 mRNA analysis in blood showed significant relationships with all corrected IQ scores in the combined cohort of FM only and PM/FM allelic groups (Table 3). A similar pattern was observed when standard IQ scores were used in blood (Figure 3a–c) and other tissues (Table S7), though some associations did not survive FDR.
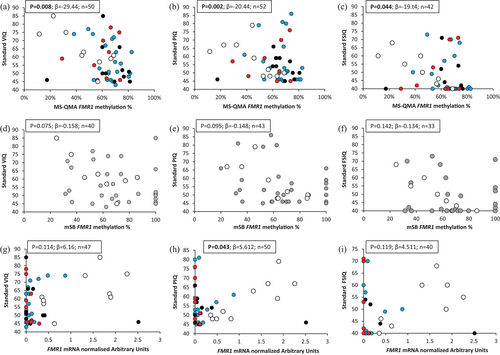
In contrast outcomes from mSB analysis of FMR1 promoter methylation in blood were not significantly associated with any of the intellectual functioning variables (Figure 3d–f, Tables 3 and S7). Interestingly, a number of males with MS-QMA methylation <40% and at the higher end of the distribution of intellectual functioning in this cohort (standard scores PIQ, VIQ, and FSIQ ≥60) were 100% methylated by mSB. However, FMR1 mRNA levels in blood did show a significant relationship with standard PIQ scores, while relationships for standard VIQ and FSIQ scores did not reach significance (Figure 3g–i).
Analysis of a sub-sample of participants from the combined FXS group (FM-only and PM/FM allelic groups combined) who had complete molecular and intellectual functioning data, demonstrated that MS-QMA MR in blood and saliva, as well as FMR1 mRNA levels in blood were significantly associated with cFSIQ and cPIQ scores (Table S8). Although these same molecular variables were associated with cVIQ (to a lesser degree) in the smaller sample size of 17 participants, these relationships did not survive FDR.
4 DISCUSSION
This study identified significant heterogeneity in FMR1 promoter methylation levels using MS-QMA in different tissues and in FMR1 mRNA levels in blood. However, this heterogeneity was not fully captured by mSB in blood from a large cohort of males affected with FXS. This “additional” variability in methylation levels detected by MS-QMA showed strong relationships between different tissues with FMR1 mRNA levels in blood and intellectual functioning. Together, these relationships demonstrate the biological and clinical significance of the variability identified by MS-QMA, that was not detected by mSB.
These findings are important because current guidelines for FXS testing (Biancalana et al., 2015) and estimates relating to mosaicism are based on the literature from the 1990s (Nolin et al., 1994; Rousseau et al., 1991a), when largely there was no alternative available to mSB, and the field of FMR1 epigenetics was in its infancy. Update of these guidelines should be considered based on more recent technological advances supported by findings from this study. One potential reason for the continued use of mSB is that all standard and long-range PCR based methods including AmplideX and FastFrax would be affected by the loss of primer binding sites that may be associated with somatic retraction. Subsequently, expanded alleles may be missed or not amplified, as demonstrated in a recent case study of two brothers with FXS (Field et al., 2019); however, mSB is not affected by this mechanism. The current study also suggests that in most instances FXS is not caused by complete silencing, but rather the decrease of FMR1 mRNA levels associated with methylation mosaicism and consequent decrease in intellectual functioning.
4.1 Re-defining mosaicism in males affected with FXS
In this study, 62.2% of males with only FM alleles showed methylation of <100% by mSB. This was higher than the previous mosaicism estimates of 12% (Rousseau et al., 1991a) to 41% (Nolin et al., 1994) reported in FM males using mSB. Although the laboratory method for mSB was similar to that used in previous studies, in this study a more sensitive automated mSB analysis protocol was utilized (Image Quant TL-software densitometry scanning and calling of bands produced by Typhoon Trio) as compared to manual calling for presence or absence of methylated bands from X-ray films used in previous studies (Nolin et al., 1994; Rousseau et al., 1991a). This could potentially explain why mSB analysis in this study reported a higher prevalence of methylation mosaicism (defined as methylation <100%) in males with FXS than in previous studies (Nolin et al., 1994; Rousseau et al., 1991a).
Importantly, in this study ~60% of males with only FM alleles who were fully methylated by mSB, also had detectable FMR1 mRNA in blood. The high proportion of males with FXS expressing detectable FMR1 mRNA, is discordant with previous literature (Nolin et al., 1994; Rousseau et al., 1991a). All of these males only had a single “methylated” FM band according to mSB, and thus should have had complete silencing of FMR1. In contrast, presence of methylation mosaicism was detected in all males with FM using MS-QMA. These MS-QMA results are consistent with the FMR1 mRNA levels and the FXS phenotype being quite heterogeneous in these “fully methylated” FM males, as reflected by variability in intellectual functioning.
4.2 Difference in location and number of CpG sites analyzed: Does this matter?
We have previously demonstrated significant relationships between MS-QMA analysis of the FMR1 exon 1/intron 1 boundary (also known as the FREE2 region) and methylation values from AmplideX mPCR (Hensel et al., 2019). The restriction sites analyzed by AmplideX mPCR are located on either side of the CGG expansion, which includes the “classical” CpG island, but are different to the sites analyzed by mSB. The weaker relationships between mSB analysis with FMR1 mRNA levels described in this study, and lower analytical sensitivity (ability to detect unmethylated alleles on methylated allele background) as compared to MS-QMA, may indicate that the methylation of the few restriction sites analyzed by mSB does not as accurately represent variability in methylation patterns between: (i) CpG sites of the whole promoter region including FREE2 (Godler et al., 2010; Stoger et al., 2011); and (ii) different cells and tissues analyzed. Because MS-QMA examines methylation of 11 CpG sites over a much larger region, it may be more representative of biologically and clinically significant changes in DNA methylation of the promoter (reviewed in [Kraan et al., 2019]), including CpG sites analyzed by AmplideX mPCR (Hensel et al., 2019). This would translate into higher analytical sensitivity. Technical differences between restriction enzyme based analysis used by mSB and bisulfite-based analyses employed by MS-QMA, each with their own limitations, as discussed in our earlier study (reviewed in [Godler & Amor, 2019]), may also explain the differences in analytical sensitivity observed in this study.
4.3 Inter-tissue mosaicism and epigenotype–phenotype relationships
Interestingly, MS-QMA methylation in saliva was more closely associated with methylation in BEC, than blood. This suggested that saliva DNA isolated in this study was primarily from cells of epithelial rather than of hematopoietic origin. Despite this inter-tissue variability, MS-QMA methylation analysis in all tissues showed significant relationships with most intellectual functioning scores. In contrast, no associations were observed between mSB methylation in blood and intellectual functioning scores (corrected or standard), with the only significant relationships observed for mSB methylation with FMR1 mRNA levels in the PM/FM allelic group. Overall, methylation determined by MS-QMA across all tissues showed similar strength as predictors of intellectual functioning. This was consistent with the analyses of relationships in the subgroup of participants who provided all biological samples across all three tissues and who also had complete mSB methylation data available.
The lack of association between mSB and intellectual functioning scores is in contrast with previous mSB studies examining relationships with the phenotype (Godler et al., 2010; Taylor et al., 1994). Specifically, relationships between severity of FXS with FMRP expression and methylation of several restriction sites within FMR1 CpG island in blood, primarily in partially methylated “high functioning” males, has previously been shown (Godler et al., 2010). In this study, however, no “high functioning” males with completely unmethylated FMR1 promoter were included in the studied FXS cohort, which may have skewed the relationships in these earlier studies.
A weak correlation has been previously reported between FMR1 promoter methylation within the CpG island examined using mSB and intellectual functioning in females with FXS (Taylor et al., 1994). In these earlier studies it has been proposed that the FMR1 CpG island can be: (i) fully methylated in typical FXS where FMR1 transcription would be completely silenced; (ii) partially methylated in mosaic cases where there would be partial transcription of FMR1 and milder phenotype than in fully methylated individuals (de Vries et al., 1996; Rousseau et al., 1994); or (iii) unmethylated with FMR1 transcription being close to normal or slightly increased levels and the FXS phenotype being least severe of all FXS groups (de Esch et al., 2014). These explanations of FXS etiology and epi-genotype–phenotype relationships, however, may be oversimplified. Evidence for this comes from a recent study in which males with FM-only CGG expansions (aged <19 years) expressing FM FMR1 mRNA had significantly more severe autism features based on the Autism Diagnostic Observation Schedule—Second Edition, compared to males with FM-only alleles who had completely silenced FMR1, despite no significant differences being observed on intellectual functioning (Baker et al., 2019). It is plausible that a FXS pathomechanism exists whereby active unmethylated FM and/or PM alleles lead to expression of toxic expanded FMR1 mRNA in some cells (not detected through methylation analysis), in conjunction with possible reduced FMR1 mRNA and FMRP levels in other cells detected using FMR1 methylation analyses.
4.4 Limitations
A limitation of this study is that all tissues examined for methylation and FMR1 mRNA levels were peripheral, while the FXS intellectual functioning phenotypes are thought to be caused largely by changes in the brain. However, significant associations between MS-QMA analysis of DNA methylation in all three tissues examined and the degree of intellectual functioning in this, and previous studies (Arpone et al., 2018; Inaba et al., 2014), suggest that exon 1/intron 1 methylation examined by MS-QMA is relatively conserved between peripheral tissues and the brain, and is predictive of the phenotype severity in males affected with FXS. Another potential limitation is that FMRP levels were not examined. Changes in FMR1 transcription do not always reflect changes in its protein product because expanded alleles may not be translated (Budimirovic et al., 2020; Primerano et al., 2002). Another limitation may be that mSB was used as a pre-assessment test, but only for the Chilean participants. For the Australian participants Southern blot analyses did not employ methylation sensitive restriction enzymes. In both cases Southern blot was used exclusively for CGG sizing to confirm presence of PM and/or FM alleles, and not for quantitative methylation analysis. Moreover, the diagnostic pre-assessments followed different protocols to those used in this study to quantify methylation using Southern bl. Broad range in patient age and small sample size for specific age groups are other limitations which should be addressed in future studies examining the relationships between intellectual functioning with the methylation status of the FMR1 promoter.
In summary, this study demonstrates that complete silencing of FMR1 is not common in FXS, and that previous estimates of the proportion of FXS males with presence of methylation mosaicism with incomplete FMR1 silencing are underestimated (Nolin et al., 1994; Rousseau et al., 1991a). This may be potentially due to analytical sensitivity issues associated with mSB. MS-QMA in blood, saliva and BEC offered several advantages over the mSB for measurement of FMR1 promoter methylation in males with FXS, including a more refined resolution of methylation differences near the upper and lower detection ranges for low-level mosaicism, much lower DNA requirements and much higher throughput. The clinical and biological significance of this low-level mosaicism has been demonstrated through relationships between methylation changes detected by MS-QMA (either above 80% or below 20%), FMR1 mRNA levels in blood, and intellectual functioning in males affected with FXS. If confirmed in future independent studies, this will have implications for prognostic utility of methylation testing in diagnostic settings and for patient stratification in FXS clinical trials.
ACKNOWLEDGMENTS
We are grateful to study participants and their families for their contribution. We also thank family support groups including the Jonathan Cohen from the Fragile X Alliance Inc, the Fragile X Association of Australia, and Justine Elliot from Victorian Clinical Genetics Services for their assistance with participant recruitment. We would also like to thank the following individuals for their assistance with the administration and coding/scoring of neuropsychological assessments: Cherie Green, Nusrat Ahmed, Annabelle May Marsh, Jaqueline Maya, and Pura Ballester Navarro. We would also like to thank Solange Aliaga for performing Southern blot analyses utilized in this study, and Lesley Bretherton and Howard Slater for their contributions.
This work was funded by The Victorian Government's Operational Infrastructure Support Program, Murdoch Children's Research Institute, Royal Children's Hospital Foundation, Martin & E.H. Flack Trust, Pierce Armstrong Trust, Financial Markets Foundation for Children (Australia) (FMFC; grant number: 2017-361 to David E. Godler), the National Health and Medical Research Council (NHMRC project grant numbers: 1049299 and 1103389 to David E. Godler; NHMRC Early Career Fellowship project grant number: 1112934 to Claudine M. Kraan). David E. Godler salary was also supported by the Next Generation Clinical Researchers Program - Career Development Fellowship Funded by the Medical Research Future Fund (grant number 1141334). Angelica M. Alliende was funded by the Comision Nacional de Investigacion Cientifica Y Tecnologica (CONICYT) and Chile's National Commission for Scientific and Technological Research.
Marta Arpone was supported by the International Postgraduate Research Scholarship (IPRS) and the Research Training Program Scholarship funded by the Australian Government and awarded by the University of Melbourne, and in part by the Diagnosis and Development group of the Murdoch Children's Research Institute. Open access publishing facilitated by The University of Melbourne, as part of the Wiley - The University of Melbourne agreement via the Council of Australian University Librarians.
CONFLICT OF INTEREST
Dr. David E Godler is as an inventor on patents related to the technologies described in this publication and is an Executive Director of Epigenetic, Diagnostic & Genetic screening (E.D.G). Innovations & Consulting that receives funds from this intellectual property. He has also acted as a paid consultant for Bellberry, Ltd. and Actinogen Medical, Pty, Ltd. No other authors have conflict or competing interests to declare.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.




