The diagnostic utility of exome-based carrier screening in families with a positive family history
Abstract
Identification of disease-causing variants in families with a history of a suspected recessive disorder is essential for appropriate counseling and reproductive decision making. The present case series depicts the utility of whole exome-based phenotypes-driven carrier analysis in 14 families with a positive family history. A phenotype-based analysis revealed a putative diagnostic yield of 71.4%. Proband sample, though insufficient, was available in only one family, which allowed the diagnosis to be confirmed. In the remaining nine families, despite the detection of heterozygous pathogenic/likely pathogenic variants, only a putative diagnosis was possible due to incomplete proband phenotyping as well as nonavailability of proband samples. We describe the youngest known patient homozygous for a likely pathogenic variant in PPP1R21. He is currently asymptomatic at 7 days of life and has a simplified gyral pattern on neuroimaging. The case series, though small, captures the challenges in the diagnosis of genetic disorders in low to middle income countries with in-equitable health care access. It reinforces the significance of detailed phenotyping in the proband as well as the importance of DNA storage for a conclusive diagnosis. A recurring post-test counseling challenge was risk ascertainment and reproductive decision making in subsequent pregnancies if the detected pathogenic/likely pathogenic variants are co-inherited, in families with a putative diagnosis. When opted for, prenatal testing in such a scenario would be limited in its ability to comment on the fetal status with respect to the disorder in the proband.
1 INTRODUCTION
Rare genetic disorders are a hidden burden in the developing countries with an estimated 70 million individuals affected in India, in part due to consanguineous and endogamous marriages that are a common practice (Angural et al., 2020; GUaRDIAN Consortium, Sivasubbu, & Scaria, 2019). A prerequisite to appropriate reproductive decision making in these disorders is the identification of an underlying genetic etiology. A diagnosis is molecularly confirmed upon the detection of pathogenic/likely pathogenic (P/LP) variants consistent with both the inheritance pattern as well as clinical phenotype in the proband (Richards et al., 2015). Factors affecting this diagnostic odyssey include early mortality, access to health care facilities, and cost of testing (Aggarwal & Phadke, 2015; Bansal et al., 2020).
Next generation sequencing (NGS) which allows the simultaneous assessment of multiple genes has been utilized for carrier screening and reproductive decision making (Bell et al., 2011; Stals et al., 2018). Population-based carrier screening for recessive disorders began with targeted screening for disorders higher in carrier frequency and specific to ethnic groups (King & Klugman, 2018; Mastantuoni et al., 2018). A carrier frequency of 1 in 100 or more is one among the recommended criteria for disease inclusion within expanded carrier screening. This may not be suitable for individuals with a positive family history, as all genes implicated for a phenotype may not fulfill the above criteria (ACOG Committee Opinion, 2017). The ability to utilize parental exome sequencing to diagnose familial rare disorders where insufficient DNA of the proband was available has been demonstrated (Stals et al., 2018). In the present article, we present a case series wherein partner/s with a previous affected child underwent whole exome-based analysis for carrier status of suspected familial recessive disorder and discuss the caveats involved in correlating the results when the phenotype is limited and where DNA of the proband is unavailable.
2 MATERIALS AND METHODS
2.1 Participants
Inclusion criteria for the series included couples who had undergone genetic testing at Neuberg Centre for Genomic Medicine (NCGM), Ahmedabad, during the period between January 2019 and December 2020, for carrier status analysis, based on positive family history due to previously affected child/children (Table 1). All available data were retrospectively reviewed, with the approval of the NCGM Institutional Review Board. Availability of proband sample for analysis was sought for, which was available in only one family (Family 1). Extensive pretest counseling was performed, especially emphasizing the uncertainty of the carrier-alone approach to confirm a retrospective diagnosis in absence of a proband sample. Couples evaluated as a part of routine preconception screening (absence of family history) were excluded. Individuals who underwent non-NGS-based carrier status evaluation through genetic testing other than exomes (such as MLPA, PCR, Sanger sequencing, etc.) were excluded. A total of 14 families, out of which 9 reported consanguinity, met inclusion criteria for exome-based, phenotype-driven carrier screening and are reported here. After informed consent, eight couples had opted for simultaneous analysis while sequential screening was pursued in remaining cases. For the couples who opted for sequential analysis, subsequent partner testing was performed based on informed consent after posttest counseling via whole exome/targeted gene panel/targeted variant analysis. Post-test genetic counseling was provided to all couples to help them understand the significance as well as limitations of the genetic testing results with respect to a confirmed diagnosis in the deceased proband, in the absence of proband sample availability.
| Family ID | Clinical features | Consanguinity | Test requested | Gene | Variant details (classification) | Results – female partner | Results – male partner | OMIM phenotype | Follow-up |
|---|---|---|---|---|---|---|---|---|---|
| Group A: Confirmed diagnosis with respect to proband phenotype | |||||||||
| 1 | First affected pregnancy terminated due to multiple anomalies: unilateral club hand, horseshoe kidney, corpus callosum agenesis. Microarray normal | Yes | Couple exome | PALB2 | NM_024675.4:c.2716delT p.Trp906Glyfs*17 (LP) |
Heterozygous | Heterozygous | Fanconi anemia complementation Group N (MIM#610832) Breast cancer susceptibility (MIM#114480) |
CVS-fetus homozygous normal. Child healthy 6 months old male |
| Group B: Putative diagnosis due to strong concordance with proband phenotype | |||||||||
| 2 | One affected male succumbed to suspected IEM. Tandem mass spectrometry: elevated C3 acylcarnitine and total carnitine. No urine gass chromatogrpahy performed. Filter paper could not be retrieved. Clinical suspicion: propionic acidemia, methylmalonic acidemia | No | Couple exome | PCCB* | NM_000532.5:c.183+5G>A (P) |
Heterozygous | Heterozygous | Propionic acidemia (MIM#606054) | CVS-fetus homozygous normal. A healthy female currently 7 months of age. Blood TMS and urine GCMS – normal (performed on D-7 of life) |
| 3 | One affected female with blisters noted at birth over arms, legs and neck, oral epithelisosis and feeding difficulties with recurrent regurgitation. Clinical suspicion: epidermolysis bullosa with pyloric stenosis | Yes | Couple exome | ITGB4* | NM_000213.5:c.600dupC p.Phe201Leufs*15 (P) |
Heterozygous | Heterozygous | Epidermolysis bullosa, junctional, with pyloric atresia (MIM#226730) | |
| 4 | Two affected children with infantile onset nephrotic syndrome. Urine protein: 376.8 mg/dL, S.cholesterol-334 mg/dL (<200 mg/dL), S.Albumin: 14 gm/L (35–50 gm/L). Ultrasound: bilateral renal parenchymal change with moderate fluid collection in the peritoneal cavity | NA | Exome sequencing in female partner. Nephrotic syndrome gene panel in male partner | PLCE1 | NM_016341.3:c.4483C>T p.Arg1495Ter (LP) |
Heterozygous | Heterozygous | Nephrotic syndrome, type 3 (MIM#610725) | |
| Group C: Inconclusive diagnosis due to partial concordance to proband phenotype | |||||||||
| 5 | One affected child with occulo-cutaneous albinism. Succumbed to meningo-encephalitis at 1.6 years | Yes | Couple exome | BLOC1S6 | NM_012388.4:c.224+1G>A (LP) |
Heterozygous | Heterozygous | Hermansky-Pudlak syndrome 9 (MIM#614171) | PGT-M. Lost to follow-up |
| 6 | Two affected females. First female succumbed to prematurity and neonatal sepsis. Second female succumbed at 8 months. Features: hypotonia, global developmental delay and repeated aspiration. Neuroimaging – not performed | Yes | Sequential exome | PPP1R21 | NM_001135629.3:c.763delA p.Ile255Leufs*13 (LP) |
Heterozygous | Heterozygous | Neurodevelopmental disorder with hypotonia, facial dysmorphism, and brain abnormalities (MIM#619383) | CVS in third conception-fetus homozygous affected. Pregnancy terminated. Fetal autopsy and imaging declined. CVS (fourth conception): affected homozygous. Delivered male currently 7 days old, asymptomatic. Neuroimaging (Day 2 of life): simplified gyral pattern with prominent ventricles |
| 7 | Two affected fetuses with hydrocephalus and corpus callosum agenesis on antenatal ultrasounds. Fetal karyotyping –normal. No fetal DNA available | Yes | Exome in female partner. Targeted variant analysis in male partner | TRAPPC12* | NM_016030.5:c.361G>T p.Glu121Ter (LP) |
Heterozygous | Heterozygous | Encephalopathy, progressive, early-onset, with brain atrophy and spasticity (MIM#614139) | |
| 8 | Two affected males. Death in the neonatal period due to respiratory failure. Hypotonia with absent reflexes noted. Neuroimaging – normal. Tandem mass spectrometry – normal | No | Couple exome | LMOD3 | NM_198271.5:c.235A>T p.Lys79Ter (LP) |
Heterozygous | Heterozygous | Nemaline myopathy 10 (MIM#616165) | |
| 9 | Three affected pregnancies with non-immune hydrops fetalis. Karyotyping in the second was normal | No | Sequential exome | HEXA* | NM_000520.5:c.1385A>T p.Glu462Val (P) |
Heterozygous | Heterozygous | Tay-Sachs disease (MIM#272800) | |
| 10 | Two affected (one male, one female). Non-immune hydrops fetails with polyhydramnios. Karyotyping normal Death within 24 h due to respiratory failure. Couple karyotying normal | Yes | Couple exome | GBE1 | NM_000158.4:c.321G>A p.Trp107Ter (LP) |
Heterozygous | Heterozygous | Glycogen storage disease IV (MIM#232500) | |
| Group D: No significant phenotype related variants detected | |||||||||
| 11 | One female succumbed at Day 10 of life. No antenatal complications. No birth asphyxia. Intractable seizures on Day 2 of life with poor feeding. Medical records unavailable for review. No neuroimaging done | Yes | Couple exome | GPHN | NM_020806.4:c.828+1G>A (LP) | No variant detected | Heterozygous | ||
| 12 | Two pregnancies terminated due to arthrogryposis multiplex congenita. Fetal karyotyping in first conception normal | Yes | Couple exome | Negative | |||||
| 13 | Previous pregnancy terminated due to fetal anomalies. Bilateral enlarged cystic kidneys and meningomyelocele. No fetal testing performed. Couple karyotype normal | No | Exome sequencing in male partner only | Negative | |||||
| 14 | First born with imperforate anus (operated), ventricular septal defect and intractable spasms. EEG – s/o hypsarryhtmia. Neuroimaging – normal. Karyotype –normal. Couple karyotyping – normal | No | Couple exome | Negative | |||||
- Note: Corresponding variant (*) reported in clinvar database.
- Abbreviations: CVS, chorionic villus sampling; IEM, inborn errors of metabolism; LP/P, likely pathogenic/pathogenic; PGT-M, preimplantation genetic testing for monogenic disorders.
2.2 Methodology
Included partner/s underwent whole exome sequencing as per standard protocols. Briefly, genomic DNA was extracted, and libraries were prepared using a custom capture kit. Paired end sequencing was performed with 2 × 100/2 × 150 chemistry using NovaSeq 6000 (Illumina Inc., San Diego, CA) at a mean depth of 100-150X and a >95% coverage at 20X. Sequenced reads were assembled and aligned to reference sequences based on NCBI RefSeq transcripts and human genome build GRCh37/UCSC hg19.
2.3 Variant inclusion and classification
Analysis was focused on the phenotype present in the proband which was obtained through history and examination of previous medical records. Variant filtering was performed via the following: (1) Human phenotype ontology (HPO) terms relevant to the clinical presentation in the proband. (2) Variants in genes common to both partners were filtered and analyzed for phenotype consistency with the proband. (3) Rare variants with a MAF of <0.01% were filtered and analyzed for phenotype consistency with the proband. Only variants with a minimum read depth of 10X and Phred score >30 were included in the analysis. Variants were classified according to the 2015 American College of Medical Genetics (ACMG) sequence variant interpretation criteria (Richards et al., 2015). In accordance with carrier screening, reporting was mainly restricted to variants classified as pathogenic/likely pathogenic (P/LP) as per ACMG guidelines. All reported variants were Sanger confirmed. At the time of the above analysis, copy number variant (CNV) calling was not integrated with whole exome analysis. Incidental findings (Kalia et al., 2017) were reported when requested.
3 RESULTS
Eleven families underwent preconception NGS screening, and three families were evaluated during an ongoing pregnancy. DNA of the affected was available in only one family (Family 1) and was utilized for confirmatory testing due to insufficient amount. The extent of preliminary investigations performed in each respective proband varied among the cases, with six probands having a nonspecific heterogeneous phenotype and insufficient preliminary work up. P/LP variants with phenotypic overlap were observed in 11 families; however, a robust genotype–phenotype correlation was only possible in four. Variants concordant with phenotype were not identified in two cases. In two consanguineous couples with suspected IEM and duo-analysis, an LP variant was identified in one partner only, hence was nondiagnostic (Table 1). Prenatal testing for the detected variants was performed for three families while pre-implantation genetic diagnosis (PGT-M) was performed in one. Information pertaining to clinical features, test performed, detected variants, and relevance to observed phenotype for 14 cases is provided in Table 1. Select families (Family 1, 6, and 7) exemplifying the caveats in carrier-only analysis and the nuances of genetic counseling have been described below.
3.1 Family 1
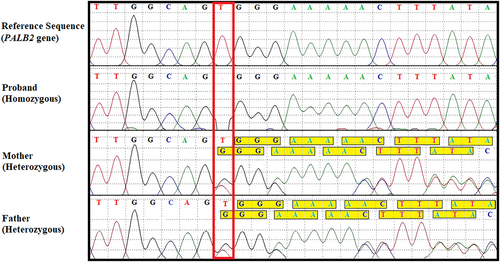
A consanguineous couple with a previous pregnancy which had been terminated due to multiple anomalies. The features included subcutaneous edema, partially unossified nasal bone, low set ears, micrognathia, right sided club hand (absence of radius), horseshoe kidney, and agenesis of corpus callosum. A microarray had been performed and was normal. Limited DNA of the affected fetus was available and hence a couple carrier screen was opted for. This revealed a heterozygous previously reported frameshift variant (p.Trp906Glyfs*17) in PALB2 implicated in Fanconi anemia, complementation group N, when inherited in a recessive manner. Fanconi anemia is associated with limb defects including radial club hand (Sevilla-Montoya et al., 2017). Corpus callosum anomalies including partial agenesis have been reported previously in patients with Fanconi anemia (Stivaros et al., 2015). Sanger analysis on stored fetal DNA detected the variant to be homozygous thus confirming the diagnosis of Fanconi anemia in the same (Figure 1). Disease-causing variants in PALB2 in recessive form lead to Fanconi anemia while in dominant state PALB2 variants lead to increased risk of cancer (Janatova et al., 2013).
3.2 Family 6
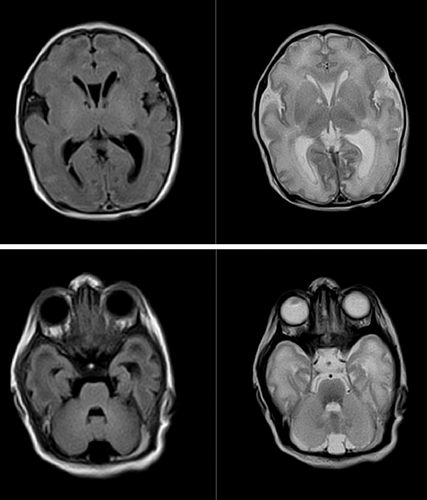
A consanguineous couple of Indian descent was evaluated at 8 weeks of gestation with respect to the history of untimely demise of two children. The first child was a prematurely delivered girl (28 weeks' gestation) who succumbed to sepsis within 1 week. The second female child succumbed at 8 months. She was born at term with a birth weight of 5.51 pounds. There is no history suggestive of birth asphyxia. Feeding difficulties in the form of poor sucking had been noted due to which she had been bottle-fed. At 1.5 months of age she was treated for aspiration pneumonia. She was readmitted at 3 months and 5 months for aspiration and chest infection when profound developmental delay and hypotonia were noted. She succumbed to aspiration pneumonia at 8 months before a specialist opinion or investigations into the etiology were possible. She had attained partial neck holding, could not recognize her parents, was unresponsive to sound and had poor visual following at 8 months. The parents desired prenatal testing. A possible autosomal recessive neuromuscular disorder was suspected based on clinical profile and presence of consanguinity. The limitations of the poorly defined phenotype in variant prioritization and phenotype-correlation as well as the inability to confirm the diagnosis in the absence of proband DNA were informed. The couple opted for sequential whole exome. A phenotype-based analysis on the husband revealed two variants: (a) a novel frameshift variant (p.Ile255Leufs*13) in the PPP1R21 gene. (b) A rare novel missense variant (p.Gly174Trp) in the MED25 gene. Biallelic loss of function variants in PPP1R21 have been reported with a rare neurodevelopmental syndrome characterized by developmental delay, hypotonia, recurrent chest infections, facial dysmorphism and variable neuroimaging abnormalities (Rehman et al., 2019). Homozygous MED25 variants have been reported with growth retardation, developmental delay, neuroimaging abnormalities along with ophthalmological and cardiac involvement (Basel-Vanagaite et al., 2015). Subsequent spouse whole exome sequencing revealed her to be the carrier of same LP variant in PPP1R21 gene. No variants in MED25 were detected in his wife. Despite the detection of the heterozygous LP variants in the couple, an unequivocal diagnosis of PPP1R21-related disorder in the proband was not possible in the absence of neuroimaging and proband DNA. The above was communicated to the couple during post-test counseling. They opted for prenatal testing despite the above limitations. A chorionic villus sampling revealed the fetus to harbor a PPP1R21 variant in homozygous state. They made an informed decision to terminate the pregnancy. Fetal investigations including autopsy was denied. Prenatal testing was again availed in a subsequent conception, where the variant was detected in a homozygous state. The family opted to continue the pregnancy. Currently they have a 7-day old male who was delivered at term with a birth weight of 6.17 pounds and an occipital-frontal circumference (OFC) of 34 cm. He was admitted for tachypnea, transient neonatal. Neurological examination was normal at birth except for incomplete Moro's reflex. Other features include retrognathia, bilateral undescended testis, and bilateral deep vertical plantar creases. No gross dysmorphic features or feeding difficulties are present. Neuroimaging on Day 2 of life revealed a simplified gyral pattern with mild ventricular prominence (Figure 2). The cerebellum and midbrain are normal. Pachygyria as well as prominent ventricles have been described in affected patients, though no data are available on imaging findings immediately after birth (Loddo et al., 2020). Further follow-up will be required to confirm the diagnosis (Figure S1, Supporting Information).
3.3 Family 7
A consanguineously married 27-year-old female of Turkish origin had two previous fetal deaths (one male and one female) due to hydrocephalus, corpus callosum agenesis. A fetal karyotype had been performed in the first pregnancy and was normal. Couple karyotyping was normal. Consanguinity and recurrence suggest a probable autosomal recessive disorder. A phenotype-driven carrier analysis was requested for the female partner by the referring clinician. The analysis revealed a heterozygous termination (p.Glu121Ter) in the TRAPPC12 gene which has been reported as likely pathogenic variant in ClinVar database. Homozygous or compound heterozygous mutations in this gene are known to cause early onset progressive encephalopathy with brain atrophy and spasticity. Antenatal detection of corpus callosum agenesis has been reported previously in one patient where postnatal imaging had confirmed the same along with pontine hypoplasia (Milev et al., 2017). Sanger analysis of the husband revealed him to be heterozygous for the same variant. The possibility of other disorders pertaining to corpus callosum agenesis was discussed and an exome-based evaluation for the husband was recommended for the same. They were informed that even in the event of such an analysis, a conclusive diagnosis of “what happened” in their previous conceptions would not be possible due to the absence of fetal DNA. They declined further testing. The couple were counseled that in the absence of a confirmed diagnosis, prenatal testing with respect to familial disorder should be considered with caution. The need to follow subsequent pregnancies via fetal ultrasound and neuroimaging was discussed. The need for fetal autopsy and extended genetic analysis to exclude other phenotypic overlaps in the event of recurrence was also discussed.
4 DISCUSSION
Couples with a positive history of a genetic disorder often opt for prenatal/preimplantation genetic testing to avoid recurrence (Poulton et al., 2018). In order to facilitate appropriate counseling regarding reproductive decisions, a confirmed molecular diagnosis in the proband with respect to the underlying phenotype is undoubtedly quite important. High morbidity and mortality, along with the need for extensive investigations, often preclude a diagnosis and culminates in a scenario where neither the proband nor DNA is available for genetic evaluation (Wojcik et al., 2019). Carrier screening with respect to the suspected familial genetic disorder would be incomplete and inconclusive if proband DNA is unavailable. Our case series, though small, captures the challenges in the diagnosis of genetic disorders in low to middle income countries with in-equitable health care access (Thong et al., 2018). It specifically depicts the challenges and ethical concerns which arise when carrier screening is opted for by families with a previous affected child in the absence of proband DNA and limited phenotyping. The availability of carrier screening was discussed by their respective primary health care provider and interested couples were referred for pretest counseling. All families where proband DNA was unavailable were duly informed of the challenges in confirming the diagnosis of the suspected familial disorder.
A confirmed diagnosis could only be made in Family 1 where stored fetal DNA was available. The observed likely pathogenic variant in PALB2 would lead to 25% risk for the fetus to be affected with Fanconi anemia, 50% chance of inheriting a cancer susceptible allele and a 25% chance of being wild-type homozygous in subsequent conceptions. Since PALB2 is associated with increased cancer susceptibility in heterozygous state, the couple were advised screening according to NCCN guidelines with respect to cancer susceptibility (Daly et al., 2020). Detailed family history revealed the maternal sister to be having Ewing's sarcoma. This has been reported earlier in only one family (Gururangan et al., 2016). Though the affected sister declined testing, there is a high likelihood that Ewing's sarcoma was related to PALB2 variants.
A diagnosis was confirmed in one family (Family 1, Group A) while only a putative diagnosis was possible in three families (Group B) where there was strong phenotypic concordance with respect to the clinical phenotype in the proband based on robust phenotyping and ancillary testing. More importantly, a subgroup of families existed (n = 6, Group C) where a conclusive diagnosis with respect to the phenotype in the proband was not possible due to nonspecific phenotypes and incomplete or negligible preliminary work-up. Family 6 exemplifies the need for detailed phenotyping including ancillary investigations like neuroimaging and genetic testing on the proband. Though neuroimaging has been performed on their third born male, who is homozygous for the likely pathogenic PPP1R21 variant, confirmation of the diagnosis with respect to that described in literature will require further follow-up.
In Family 7 where TRAPPC12 variants were detected, phenotypic concordance could not be established as corpus callosum agenesis as well as ventriculomegaly are observed in multiple genetic disorders (She et al., 2021).
Apart from the above two described families, a conclusive diagnosis could not be made in three other families within the same subgroup. Family 8, where heterozygous LP variants were detected in LMOD3, had lost two males to neonatal respiratory failure. The antenatal period had been uneventful; both children were delivered at term but required immediate ventilator support due to inadequate respiratory efforts and succumbed within 48–72 h. Tandem mass spectrometry (TMS) had been performed in the second child but efforts to retrieve the filter paper were unsuccessful. Neonatal respiratory failure has been described with LMOD3-related nemaline myopathy but is not unique to the same disease (Yuen et al., 2014).
The remaining two unrelated families (Families 9 and 10) were evaluated for recurrent nonimmune hydrops fetalis (NIH). The LP variant detected in heterozygous state in both partners in Family 9 in HEXA is interesting. The HEXA (p.Glu462Val) variant is a Founder variant within the Parmar community to which the couple belonged (Mistri et al., 2014). Lysosomal storage disorders are implicated in approximately 15% cases of NIH; however, HEXA variants are uncommon (Al-Kouatly et al., 2020). Hence the probability that the above detected variant could be a mere coincidence with respect to their ethnicity could not be overruled. In Family 10, a heterozygous LP variant was detected in GBE1 in both. A fatal perinatal neuromuscular phenotype leading to nonimmune hydrops has been reported with GBE1 but the same could not be tested on any of the affected (Zhou et al., 2021).
The availability of fetal tissue enabled a confirmed diagnosis in Family 1. In Families 2–5 detailed phenotyping +/− preliminary investigations on the proband led to the detected variants being designated as having a strong phenotypic concordance, hence enabling reproductive decision making with respect to the suspected familial genetic disorder. Negative results in the remaining families could be due to the following: (a) Presence of a copy number variant, as calling for the same on exome data had not been integrated at the time of the above analysis; (b) presence of disease-causing variants in difficult to sequence regions; (c) the possibility of a novel yet to be recognized disorder; (d) de novo variant in deceased proband.
Interpreting parental results with limited/ incomplete proband phenotyping, is challenging raising ethical concerns with respect to prenatal testing as the suspected familial disorder could not be confirmed. Though analysis and reporting was limited to pathogenic and likely pathogenic variants with some concordance to proband phenotype, a conclusive diagnosis could not be obtained in the majority, underscoring the significance of detailed phenotyping in the proband as well as the importance of DNA storage. Should these families be offered prenatal testing in the absence of a confirmed diagnosis? While all these families were duly informed of the inability to confirm the “suspected proband genetic disorder” and challenges in interpreting fetal status with respect to the same, a recurring question (irrespective of incomplete phenotype concordance) was whether the co-inheritance of the detected variants would pose a risk for the respective autosomal recessive disorder in subsequent pregnancies. Along the principles of carrier screening, since the detected variants were classified to be P/LP, their co-inheritance would be associated with a 25% risk of recurrence. An informed decision in such a scenario would have to be made by all the involved stakeholders considering the various ethical quandaries at stake.
ACKNOWLEDGMENTS
We are grateful to the patients and their families for their participation and to their physicians and genetic counselors for providing samples and clinical histories. We thank Dr. Malay Dhebar and Dr. Anand Iyer for their assistance in interpretation of neuroimaging findings.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
AUTHOR CONTRIBUTIONS
Udhaya Hardik Kotecha, Parth Shah, and Nidhi Shah designed the study and analysis. Udhaya Hardik Kotecha and Mehul Mistri performed the analysis, Udhaya Hardik Kotecha and Pranavchand Rayabarapu were involved in drafting and review of literature. All the authors read and approved the final draft.
Open Research
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.