Biallelic variants in COX4I1 associated with a novel phenotype resembling Leigh syndrome with developmental regression, intellectual disability, and seizures
Abstract
Autosomal recessive COX4I1 deficiency has been previously reported in a single individual with a homozygous pathogenic variant in COX4I1, who presented with short stature, poor weight gain, dysmorphic features, and features of Fanconi anemia. COX4I1 encodes subunit 4, isoform 1 of cytochrome c oxidase. Cytochrome c oxidase is a respiratory chain enzyme that plays an important role in mitochondrial electron transport and reduces molecular oxygen to water leading to the formation of ATP. Defective production of cytochrome c oxidase leads to a variable phenotypic spectrum ranging from isolated myopathy to Leigh syndrome. Here, we describe two siblings, born to consanguineous parents, who presented with encephalopathy, developmental regression, hypotonia, pathognomonic brain imaging findings resembling Leigh-syndrome, and a novel homozygous variant on COX4I1, expanding the known clinical phenotype associated with pathogenic variants in COX4I1.
1 INTRODUCTION
Biallelic variants in COX4I1 (OMIM: 123864) have been previously described in a patient who presented with short stature, poor weight gain, dysmorphic features and features of Fanconi anemia (Abu-Libdeh et al., 2017). COX4I1, located at 16q24.1, encodes the subunit IV isoform 1, the principal isoform for COX-IV subunit of cytochrome c oxidase (COX) or complex IV in human beings and other vertebrates. COX plays an important role in oxidative phosphorylation by transferring electrons from cytochrome c to molecular oxygen and contributes to a proton electrochemical gradient across the inner mitochondrial membrane necessary for ATP formation (Li, Park, Deng, & B, 2006). Complex IV consists of 14 different subunits, of which three are encoded by mitochondrial DNA (COX subunits I–III) and form an important catalytic core of the enzyme. The remaining 11 subunits encoded by nuclear DNA (COX subunits IV, Va, Vb, VIa, VIb, VIc, VIIa, VIIb, VIIc, VIII, and NDUFA4) are found to be tightly bound to subunits I–III (Sinkler et al., 2017). COX IV exists in two isoforms in humans and other mammals, COX subunit IV isoform 1 (COX4I1) and COX subunit IV isoform 2 (COX4I2). COX4I1 is ubiquitously expressed in mammals (Sinkler et al., 2017). Mitochondrial diseases resulting in COX deficiency (OMIM: 220110) present with marked clinical heterogeneity ranging from fatal neonatal lactic acidosis to adult myopathy. Here, we describe a novel COX4I1 variant in two siblings who present with developmental regression, seizures, and pathognomonic changes in brain imaging resembling a Leigh syndrome phenotype.
2 CLINICAL REPORT
2.1 Patient 1
Patient 1 is a 3-year-old male who was born to consanguineous Iraqi parents at 37 weeks of gestation via C-section. His birth weight was 3.316 kg (13th percentile). He had normal growth and development throughout the first 8 months of life. Developmental regression of motor skills became evident at 8 months of age when he stopped crawling and standing with support. An extensive biochemical work up, including acylcarnitine profile, plasma amino acids, urine organic acids analysis, and creatine kinase (CK), was unremarkable. Lactate was slightly elevated at 3.0 mmol/L (Reference range: 0.2–2.0 mmol/L). Brain MRI showed mild to moderate generalized cerebral/cerebellar atrophy and evidence of bilateral hypertrophic olivary degeneration with conspicuous non-enhancing lesions along the medullary pyramid and subtle signal changes along the bilateral basal ganglia and cerebellar fossa (Figure 1a). These findings were concerning for mitochondrial encephalopathy, which prompted a referral to the Genetics Clinic at Texas Children's Hospital.
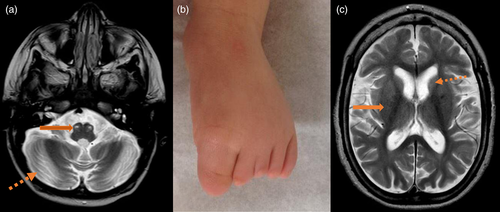
Upon evaluation at the Genetics Clinic at 13 months of age, he had further regression of his motor skills. He was unable to sit without support and did not have head control. He had profound hypotonia and required gastrostomy tube placement for feedings. Family history was significant for a 9-year-old male sibling with a similar clinical presentation (Patient 2). At the time of the initial visit, his weight was 8.82 kg (13th percentile), his height was 74 cm (8th percentile), and his head circumference was 44.5 cm (7th percentile). Physical examination did not show any dysmorphic features except bilateral hypoplasia of the 2nd–5th toes (Figure 1b). He also had axial and appendicular hypotonia and patellar hyperreflexia. He developed seizures at 2 years of age, described as epileptic spasms without hypsarrhythmia, as captured on electroencephalogram (EEG). His epilepsy is currently well controlled with zonisamide and levetiracetam. Untargeted metabolomics analysis was done on plasma and CSF, which showed elevated lactate and fumarate (Table 1). His chromosomal microarray analysis (oligonucleotide + SNP/single nucleotide polymorphism array) did not show any copy number variants but detected long contiguous stretches with absence of heterozygosity (AOH) encompassing 137 Mb in total, consistent with the history of parental consanguinity. Exome sequencing revealed a homozygous variant of uncertain significance in COX4I1 along with compound heterozygous variants of uncertain significance in MDN1 (Table 1). Following the diagnosis, evaluation by an ophthalmologist showed moderate hyperopia but no evidence of retinopathy or optic nerve atrophy. Echocardiogram has been recommended and a referral has been made, but the study has not been done yet on this patient. Coenzyme Q10 (ubiquinol) therapy was initiated at 8 mg/kg/day following confirmation of molecular diagnosis. Four months after initiation of therapy with coenzyme Q10, parents reported improvement in head control and tone.
| Clinical features | Abu-Libdeh et al. (2017) | Our cases | |
|---|---|---|---|
| Patient 1 | Patient 2 | ||
| COX4I1 genotype | Homozygous c.412G>A (p.Glu138Lys) | Homozygous c.454C>A (p.P152T) | Homozygous c.454C>A (p.P152T) |
| MDN1 genotype | − | c.4251 C>G (p.H1417Q) c.7640 C>T (p.2457L) |
c.4251 C>G (p.H1417Q) c.7640 C>T (p.2457L) |
| Genomes | hg19 (GRCh37) | hg19 (GRCh37) | hg19 (GRCh37) |
| Short stature | + | + | + |
| Weight<3rd centile | + | + | + |
| Microcephaly | + | + | + |
| Dysmorphic features | Prominent nasal bridge, fifth finger clinodactyly, frontal bossing | Bilateral hypoplasia of the 2nd–5th toes | − |
| Developmental regression | − | + | + |
| Epilepsy | − | + | + |
| MRI brain | Normal | Hypertrophic olivary degeneration, cerebellar volume loss | T2 prolongation in basal ganglia and thalami, volume loss of thalami |
| Elevated serum lactate | − | + | − |
| Chromosome breakage studies | + | − | − |
| Metabolomics profile plasma | N/A | Fumarate +2.9 Lactate +1.14 Glutarate +2.21 |
N/A |
| Metabolomics profile CSF | N/A | Lactate +3.4 | N/A |
- Note: Comparison between the first case presented by Abu-Libdeh et al. (2017) and our patients. Clinical, molecular, brain imaging and laboratory features are presented. In addition, metabolomics data for Patient 1 are also presented showing elevated fumarate and lactate in plasma and CSF samples respectively. Numerical values represent Z-scores of different metabolic analyte levels. Metabolomics analysis was performed at Baylor Genetics (www.BaylorGenetics.com; Kennedy et al., 2017; Miller et al., 2015).
2.2 Patient 2
Patient 2 is the older male sibling of Patient 1 and is 11 years old. He was born full term in Iraq. He met all developmental milestones at appropriate ages until 11 months of life. Regression of motor skills was noted around 11 months of age when he started to have frequent falls and was unable to stand up. Infantile spasms associated with hypsarrhythmia on EEG started at 12 months for which he was treated with ACTH. His epilepsy was also treated with topiramate and valproic acid. Motor regression continued after 12 months of age. Clonazepam was initiated to treat myoclonic jerks. He had a normal comprehensive work up including CK, plasma amino acids, urine organic acid analysis, carbohydrate deficient transferrin, lactate and pyruvate. MRI of the brain showed gliosis of bilateral basal ganglia, thalami, cerebellum and periventricular white matter with diffuse cerebral and cerebellar volume loss, and congenital hypoplasia of the inferior vermis (Figure 1c). His electromyogram (EMG) was normal. He had multiple genetic tests, including SCN1A deletion-duplication screening and sequencing and comprehensive epilepsy next generation sequencing panel, which were normal. A chromosomal microarray analysis was done, which showed multiple areas of AOH consistent with the history of consanguinity.
Upon confirmation of the sibling's diagnosis through exome sequencing, Patient 2 had known familial mutation testing for COX4I1 and MDN1, which identified the presence of the same variants as his sibling (Table 1). At the time of initial evaluation at the Genetics Clinic, he was 14 years of age. He had profound hypotonia with poor head control, inability to sit unsupported and was wheelchair bound. Like his brother, he also required gastrostomy tube for feedings. He had failure to thrive as evidenced by weight, height, and head circumference at 2nd, < 1st, < 2nd percentiles, respectively. He did not have any dysmorphic features including the toe abnormality detected in his sibling. He had notable hypotonia with strength greater on the left side than the right. EEG evolved into multifocal spikes with the pattern of epileptic encephalopathy. Skeletal muscle analysis after muscle biopsy at 8 years of age revealed mild Type II myofiber atrophy. A mitochondrial respiratory chain enzyme analysis on the muscle biopsy specimen showed that complex IV activity was reduced (Table 2). Following the diagnosis, he had an ophthalmology evaluation that revealed intermittent alternating exotropia with no evidence of retinopathy or optic nerve atrophy. Similar to his sibling, an echocardiogram has been recommended and a referral has been made but the echocardiogram has not been done yet. Coenzyme Q10 therapy (ubiquinol) was initiated at 8 mg/kg/day following confirmation of molecular diagnosis. Parents reported improvement in visual focus 4 months after initiation of coenzyme Q10 therapy.
| ETC activities | ETC complexes | Patient valuea (% of mean) | Control ± SDa |
|---|---|---|---|
| NADH-ferricyanide dehydrogenase | I | 96 (27, 44) | 360.4 ± 96.3 |
| NADH cytochrome c reductase | I + III | ||
| Total | 13.4 (47, 78) | 28.4 ± 6.1 | |
| Rotenone sensitive | 2.6 (30, 49) | 8.7 ± 3.9 | |
| Succinate dehydrogenase | II | 4.8 (50, 83) | 9.6 ± 3.0 |
| Succinate cytochrome c reductase | II + III | 1.3 (32, 52) | 4.2 ± 1.2 |
| Cytochrome c oxidase | IV | 6.5 (16, 27) | 40.3 ± 15.5 |
| Citrate synthase | 177 (60, 100) | 293.1 ± 66 |
- Note: Mitochondrial respiratory chain analysis done on muscle biopsy performed on Patient 2 shows reduction in multiple respiratory chain enzyme activities. Enzyme activities are normalized against citrate synthase (CS) activity when CS activity is greater than 1 SD above or below the control mean. The first figure inside the parenthesis represents the mean prior to normalization to CS activity. The second figure inside the parenthesis represents the data after normalization against CS activity. The cytochrome c oxidase activity was strikingly reduced at 16% compared to the controls, meeting Walker criteria.
- a nmol/min/mg protein.
3 RESULTS
A homozygous c.454C>A (p.P152T) variant in COX4I1 was detected by trio whole exome sequencing in Patient 1 and confirmed by familial mutation testing in Patient 2. Unaffected sister did not have this variant. Parental testing confirmed carrier status in both parents. Proline at amino acid position 152 is highly conserved across multiple vertebrate species from zebrafish to humans, and this variant is absent in public databases such as ExAC or gnomAD (Lek et al., 2016). In silico analyses for conservation suggests that this variant is evolutionarily conserved and constrained (phastCons score = 1, phyloP = 7.461). Furthermore, ensembl predictors (that combine multiple in silico algorithms as features to reach a prediction) support the variant to be likely damaging to the COX4I1 protein product (REVEL score: 0.839 [threshold >0.75 implies damaging]). The identified homozygous COX4I1 variant resides in one of the AOH blocks spanning ~5.3 Mb located on chromosome 16q23.1–16q24.1. Both Patient 1 and Patient 2 had normal chromosomal breakage studies. Exome sequencing also identified compound heterozygous variants of uncertain significance in MDN1: c.4251 C>G/p.H1417Q (inherited from father) and c.7640 C>T/p.2457L (inherited from mother). Unaffected sister carried the second variant described above in MDN1.
Untargeted metabolomics profiling on plasma and CSF in Patient 1 showed elevated fumarate (Z-score 2.9), lactate (Z-score 3.4), and glutarate (Z-score: 2.21; Table 1). The mitochondrial respiratory chain enzyme analysis done on muscle biopsy specimen of Patient 2 showed that COX activity was reduced to 16% residual activity compared to control values, meeting a major modified Walker criterion (Table 2).
4 DISCUSSION
Only recently has a human autosomal recessive disorder been linked to COX4I1. Abu-Libdeh et al. (Abu-Libdeh et al., 2017) described a novel mitochondrial disease associated with a homozygous variant in COX4I1 in a 3.5-year-old female who presented with Fanconi anemia, short stature, poor weight gain, mild dysmorphic features, and normal brain MRI without features indicative of mitochondrial disease. The siblings presented herein share some phenotypic features including short stature, failure to thrive and microcephaly (Table 1) in the presence of a novel homozygous variant (p.P152T) in COX4I1. While neither sibling had evidence of Fanconi anemia, they had additional manifestations including developmental regression, intellectual disability, epilepsy, and pathognomonic changes in brain imaging resembling Leigh syndrome that were not present in the first reported case. These additional features suggest a phenotypic expansion of COX4I1 deficiency. To our knowledge, this is just the second clinical report involving a variant in COX4I1 and the first report describing a Leigh-like syndrome association with COX4I1 deficiency.
The most common presentation of mitochondrial disease in the pediatric population is in the form of Leigh syndrome (subacute necrotizing encephalomyelopathy; Lake, Bird, Isohanni, & Paetau, 2015). The clinical features, including but not limited to developmental delay and regression, dystonia, ataxia and ophthalmoplegia, are often seen in conjunction with imaging findings such as bilateral symmetric T2 hyperintensitities in basal ganglia and/or brain stem with MR spectroscopy revealing elevated brain lactate levels (Bonfante, Koenig, Adejumo, Perinjelil, & Riascos, 2016; Cavanagh & Harding, 1994; Rahman et al., 1996; Sofou et al., 2014). Due to the locus heterogeneity associated with Leigh syndrome, multiple efforts have been made to establish a genotype–phenotype correlation (Lake, Compton, Rahman, & Thorburn, 2016; Sofou et al., 2018). Here we propose that the presence of short stature, failure to thrive, microcephaly, developmental regression, intellectual disability, epilepsy, and abnormal complex IV activity could be observed as the characteristic findings in Leigh syndrome associated with COX4I1 deficiency.
Even though both siblings described here did not have hematological features and chromosome breakage studies consistent with Fanconi anemia, other hematological disorders such as sideroblastic anemia have been well described in multisystemic mitochondrial disorders such as Pearson syndrome, MLASA syndrome (mitochondrial myopathy, lactic acidosis, and SA) and complex I deficiency associated with a hemizygous change in NDUFB11 (Falcon & Howard, 2017; Lichtenstein et al., 2016; Riley et al., 2010; Tesarova et al., 2019). However, it was not until recently that defective oxidative metabolism and mitochondrial localization along with spontaneous mitochondrial fragmentation have been described in Fanconi anemia cells (Bottega et al., 2018; Cappelli et al., 2017; Pagano, Shyamsunder, Verma, & Lyakhovich, 2014). Imbalance of NAD+/NADH in COX deficiency has been postulated as the underlying mechanism of DNA instability and increased double-stranded DNA breaks (Douiev & Saada, 2018). Further follow-up in our patients is required in order to establish whether hematological features are a consistent finding that would be associated with COX4I1 deficiency.
The first variant described by Abu-Libdeh et al in COX4I1 was found to be in a conserved residue in the transmembrane helix domain that interacts with COX I and COX II leading to decreased mRNA expression and COX activity in the patient's fibroblasts (Abu-Libdeh et al., 2017). Similarly, the variant observed in the siblings presented herein is conserved across many species, and ETC analysis on the muscle biopsy specimen from Patient 2 revealed reduction in COX activity, further supporting the pathogenicity of this variant in our patients.
Interestingly, untargeted metabolomics profiling (Kennedy et al., 2017; Miller et al., 2015) performed on Patient 1 revealed elevated lactate, fumarate and glutarate, which indicate perturbation in energy metabolism secondary to mitochondrial dysfunction further providing functional evidence for the pathogenicity of the variant identified in COX4I1. Evidence of mitochondrial dysfunction by the use of untargeted metabolomics analysis in CSF and plasma may provide functional validation for variants of unknown significance observed in nuclear genes associated with mitochondrial disease, providing semi-quantitative values for TCA cycle intermediates and altered lipid metabolism as a consequence of abnormal mitochondrial function (Esterhuizen, Van Der Westhuizen, & Louw, 2017; Shayota et al., 2019; Tam et al., 2019).
Treatment with coenzyme Q10 is well established in coenzyme Q10 deficiency (Duncan et al., 2009; Rötig et al., 2000). In disorders where mitochondrial respiratory chain is affected, coenzyme Q10, in addition to restoring electron flow in the mitochondrial respiratory chain, also acts as an antioxidant and helps reduce the oxidative stress and has been recommended despite lack of proven efficacy (Hargreaves, 2014; Parikh et al., 2014). Given these facts, coenzyme Q10 was started in both patients herein presented. The treatment has provided minor clinical improvements 4 months following initiation of therapy. We hypothesize that a therapeutic trial with coenzyme Q10 in patients with COX4I1 deficiency may help stabilize the course of the disease. Compound heterozygous variants in MDN1 (p.H1417Q inherited from father and p.P2547L inherited from mother) were also observed in the trio whole exome sequencing performed in these siblings. The heterozygous variant p.P2457L was also carried by the healthy unaffected sister. The association of pathogenic variants in MDN1 with any disease in humans has not been well established. MDN1 or midasin is an essential AAA (ATPase associated with various activities) protein that plays a critical role in the biogenesis of ribosomal large subunits and is expressed ubiquitously. Although the variants identified in our patients were observed at low frequencies in public databases (His1417Gln: 19 times and Pro2547Leu 23 times in gnomAD) and were in trans, the only report linking MDN1 to a disease is that of Sanders et al., 2012 (Sanders et al., 2012). Sanders et al. reported a de novo L3468V missense variant in MDN1 in an individual within a cohort of subjects with autism spectrum disorder. However, a dominant model in our patients is unlikely given that the unaffected sister and parents had each one of the MDN1 variants. Given the paucity of data definitively associating MDN1 to a disease and a role in mitochondria function, we think that the MDN1 variants are unlikely to contribute to the disease etiology.
In summary, the clinical features of short stature, failure to thrive, microcephaly, developmental regression, intellectual disability, epilepsy and pathognomonic finding of Leigh-like syndrome on brain imaging along with untargeted metabolomics findings and the reduced COX activity proven by muscle biopsy provide functional evidence for the pathogenicity of the variants described here and further suggest expansion of the clinical phenotype linked to autosomal recessive COX4I1 deficiency.
ACKNOWLEDGMENTS
The authors would like to thank the patients, their family and all physicians involved in their care.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
INFORMED CONSENT
Genetic analysis was performed after obtaining a written informed consent from both parents. Consent to publish clinical data of the patients was obtained from both parents of the minors.
FINANCIAL SUPPORT AND SPONSORSHIP
Fernando Scaglia receives research support from NIH, BioElectron Technologies, and Stealth Therapeutics; and is an investigator in the North American Mitochondrial Disease Consortium. Nishitha R. Pillai is supported by Sanofi Genzyme ACMGF Next Generation Training Award.




