Clinical and genetic characterization of AP4B1-associated SPG47
Abstract
The hereditary spastic paraplegias (HSPs) are a heterogeneous group of disorders characterized by degeneration of the corticospinal and spinocerebellar tracts leading to progressive spasticity. One subtype, spastic paraplegia type 47 (SPG47 or HSP-AP4B1), is due to bi-allelic loss-of-function mutations in the AP4B1 gene. AP4B1 is a subunit of the adapter protein complex 4 (AP-4), a heterotetrameric protein complex that regulates the transport of membrane proteins. Since 2011, 11 individuals from six families with AP4B1 mutations have been reported, nine of whom had homozygous mutations and were from consanguineous families. Here we report eight patients with AP4B1-associated SPG47, the majority born to non-consanguineous parents and carrying compound heterozygous mutations. Core clinical features in this cohort and previously published patients include neonatal hypotonia that progresses to spasticity, early onset developmental delay with prominent motor delay and severely impaired or absent speech development, episodes of stereotypic laughter, seizures including frequent febrile seizures, thinning of the corpus callosum, and delayed myelination/white matter loss. Given that some of the features of AP-4 deficiency overlap with those of cerebral palsy, and the discovery of the disorder in non-consanguineous populations, we believe that AP-4 deficiency may be more common than previously appreciated.
1 INTRODUCTION
The hereditary spastic paraplegias (HSP) are a heterogeneous group of diseases characterized by a progressive degeneration of the spinothalamic tracts leading to spastic para- or tetraplegia. Bi-allelic loss of function mutations in any of the four adapter protein 4 (AP-4) complex subunits lead to spastic paraplegia and intellectual disability. AP-4 belongs to a family of adaptor proteins (AP-1 through AP-5), which are evolutionarily conserved heterotetrameric protein complexes that mediate vesicle trafficking in cells (Hirst, Irving, & Borner, 2013). The AP-4 complex is composed of four subunits (β4, ϵ, µ4, σ4) and is involved in trafficking of membrane proteins from the trans-Golgi-network. Mutations in the genes for each subunit have been reported to cause complex forms of HSP (Marras et al., 2016): AP4B1 (HSP-AP4B1 or SPG47, OMIM #614066), AP4E1 (HSP-AP4E1 or SPG51, OMIM #613744), AP4M1 (HSP-AP4M1 or SPG50, OMIM #612936), and AP4S1 (HSP-AP4S1 or SPG52, OMIM #614067). Mutations in AP4B1 have been thus far reported in 11 individuals from six pedigrees, mostly consanguineous families with homozygous mutations (Abdollahpour et al., 2015; Abou Jamra et al., 2011; Bauer et al., 2012; Lamichhane, 2013; Tan et al., 2015; Tuysuz et al., 2014). Here we report a detailed clinical, imaging, and molecular characterization of eight individuals with bi-allelic mutations in AP4B1. Five of these individuals had compound heterozygous mutations, and several recurrent mutations were identified. This is the largest cohort of patients with AP4B1 mutations reported to date, and demonstrates the presence of this disorder in populations with low rates of consanguinity.
2 METHODS AND MATERIALS
Patients were recruited into the collaborating institutions’ review board and/or ethics committee–approved research protocols, and written informed consent for participation, including publishing photographs and videos, was obtained. Social media and the CureSPG47 website (www.curespg47.org) were used by some families to connect with other families and the investigators. Data were collected through a retrospective, cross-sectional review of medical records. Brain imaging was performed on all patients, and 10 MRI brain scans were available for review. Genetic testing was done using multi-gene panels or exome sequencing. Variants were annotated using the ExAC database and interrogated in silico to predict damaging effects (by calculating Combined Annotation Dependent Depletion (CADD) scores [Kircher et al., 2014]). Phenotypes of previously published individuals with AP4B1 mutations were also reviewed.
3 RESULTS
3.1 Clinical characterization
We identified eight individuals with bi-allelic AP4B1 mutations from seven unrelated families. The phenotypes of the cohort are summarized in Table 1. In our cohort, the majority of patients were female (7/8). All were born at term without significant perinatal complications. The neonatal course was complicated by hypoglycemia and periodic breathing in two patients (P2 and P3). The age at initial presentation for concerns of developmental delay was early, generally between 2 and 4 months of age, with an average age at diagnosis of 54 months (range: 26 months–12 years). Excluding two siblings, who presented for genetic evaluation at significantly older ages and were identified at ages 6 and 12 years, respectively, reduces the average age at diagnosis to 36 months.
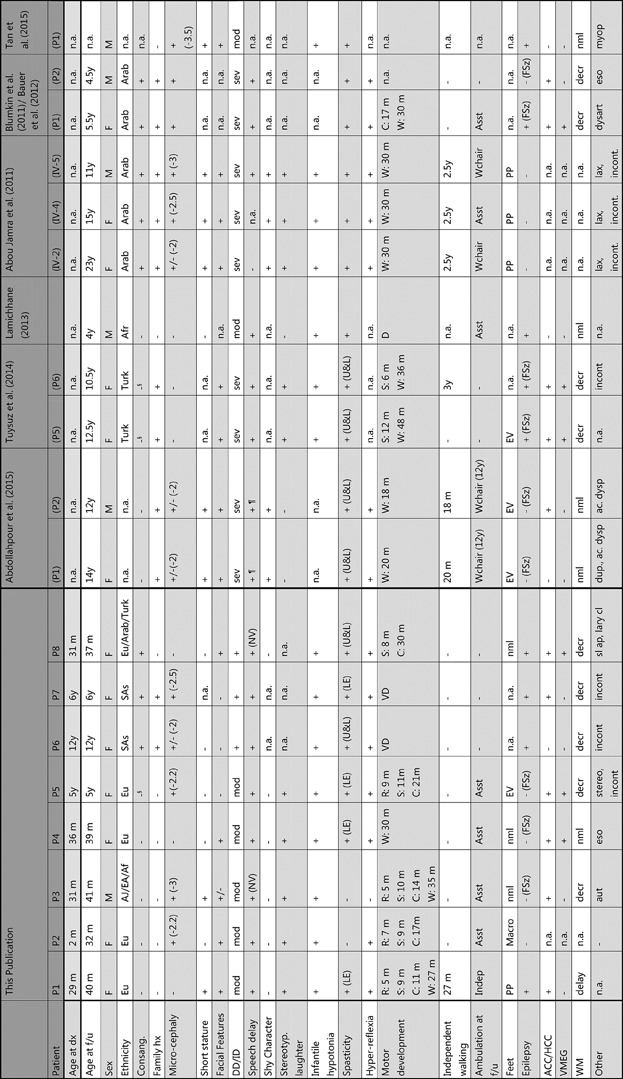
- + (feature present), − (feature absent), +/− (feature borderline), n.a. (feature not assessed). dx (diagnosis), f/u (follow up), hx (history); Ethnicity: Eu (European), AJ (Ashkenazi Jewish), EA (East Asian), Af (African), SAs (South Asian), Turk (Turkish), Arab (Arabian), § (family members from same small geographical area), Microcephaly: standard deviations below the mean shown if present; DD/ID (developmental delay/intellectual disability) is either mod (moderate, IQ 35–50), or sev (Severe, IQ 20–35), or present (+) if not specified, Speech delay: NV (non-verbal), ¶ (became non-verbal over time), Spasticity: LE (lower extremities), U&L (upper and lower extremities), Motor development: D (delayed), C (crawling), R (rolling), S (sitting), W (walking), VD (very delayed); Independent walking: age at first independent steps shown, Ambulation at f/u: Indep (independent walking), Asst (walking with assistance), Wchair (uses wheelchair, age at wheelchair onset shown), PP (pes planus), Macro (macrodactyly right 1st/2nd toe), EV (equinovarus), Epilepsy: Epilepsy defined as ≥ 2 unprovoked seizures or treatment with antiepileptic medication, FSz (febrile seizures), ACC/HCC (agenesis or hypoplasia of corpus callosum), VMEG (ventriculomegaly), WM (white matter), either delay (delayed myelination), decr (decreased), or nml (normal), Other: aut (Autism), eso (esotropia), stereo (stereotypies), incont (no sphincter control), dup (duplicated ureter), ac. dysp (acetabular dysplasia), lax (joint laxity), dysart (dysarthria), lary cl (laryngeal cleft, type 1), myop (myopia), sl ap (sleep apnea).
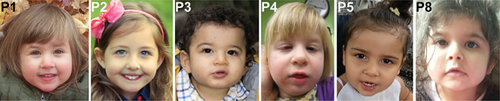
All eight patients presented with early developmental delay with prominently delayed motor milestones. Rolling was delayed to 5–7 months, independent sitting to 8–11 months, crawling to 11–30 months, and walking, if achieved and generally with assistance, to 27–35 months. Truncal and appendicular hypotonia was universally noted in early infancy and progressed to lower extremity spasticity, leading to spastic paraplegia and impaired ambulation. Several individuals were thought to have cerebral palsy prior to genetic diagnosis. The oldest individual in our cohort progressed to spastic tetraplegia with more prominent involvement of the lower extremities (Table 1). Generalized dystonic movements were described in one patient (P7). In patients who continued to walk with assistance, a wide-based, unsteady spastic gait was described. Speech development was delayed in all children and remained limited to a few words with two individuals remaining non-verbal. Microcephaly (head circumference decreased by ≥2 SD) was reported in five patients, but the range was mild (−2 to −3 SD), with three patients being normocephalic. Height and weight were normal in the majority of patients. Facial features seen in more than one individual include prominent cheeks, a thin upper vermilion, and a flat nasal bridge, but these features do not appear unique enough to make this a recognizable syndrome (Figure 1). Unprovoked seizures qualifying for a diagnosis of epilepsy occurred in four children with onset within the first 2 years of life. Febrile seizures were also common and occurred in three additional patients. Of note, the “shy character” previously described in two families (Abdollahpour et al., 2015; Abou Jamra et al., 2011) was not present. All eight children in our cohort were described as social and with a generally happy demeanor. Characteristic and stereotypic episodes of laughter were independently reported in at least five individuals (Supplemental Video S1). All patients received supportive care including early intervention, physical, occupational, and speech therapy.
In addition to reporting phenotypes of eight new patients, we reviewed the literature for previous reports of individuals with AP4B1 mutations, which are included in Table 1. Including the patients identified in this paper, a total of 19 individuals with AP4B1 mutations have now been identified, and a core set of clinical features has emerged (Table 2). Manifestations shared by the majority of individuals include: early developmental delay and intellectual disability (19/19, 100%) with prominent motor (17/17, 100%), and speech (16/17, 94%) delays, neonatal or infantile hypotonia (15/15, 100%) progressing to spastic diplegia (17/19, 89%) with loss or failure to achieve independent walking (15/17, 88%). Mild microcephaly (−2 to −3 SD range) and short stature are found in 69 and 57% of individuals, respectively. Peculiar episodes of stereotypic laughter, first video-documented here (Supplemental Video S1), were found in about two thirds of patients. Epilepsy, defined as more than two unprovoked seizures, occurred in about half the patients. Febrile seizures were also common, occurring in an additional six patients.
| Early developmental delay and intellectual disability | 100% (19/19) |
| Delayed motor development | 100% (17/17) |
| Neonatal or infantile hypotonia | 100% (15/15) |
| Delayed speech development | 94% (16/17) |
| Progression to spastic diplegia | 89% (17/19) |
| Loss of independent walking | 88% (15/17) |
| Episodes of stereotypic laughter | 77% (10/13) |
| Microcephaly (≥ − 2SD) | 69% (13/19) |
| Short stature | 57% (8/14) |
| Epilepsy | 47% (9/19) |
| Thin corpus callosum | 73% (11/15) |
| Delayed myelination or white matter loss | 67% (10/15) |
| Ventriculomegaly | 40% (6/15) |
Spastic paraplegia has not yet developed in two young patients (P2 and P3, aged 32 and 31 months, respectively), although an abnormal, unsteady gait was noted. Progression to tetraplegia was documented in the two oldest patients in our cohort (P6 and P8) and at least four previously reported patients who became non-ambulatory after the age of 10 years.
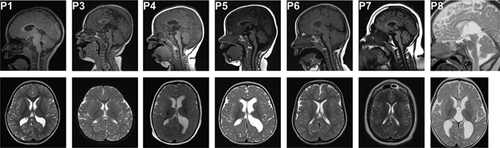
3.1.1 Brain imaging
Brain imaging is summarized in Table 1 and illustrated in Figure 2. Serial studies were available in three patients (P1, P4, and P5). Delayed myelination and/or white matter loss in the periventricular and other supratentorial regions was present in all patients but one (P4). A thin corpus callosum particularly affecting the posterior aspects was noted in 6/7 patients. Ventriculomegaly was seen in three patients, mostly in the form of prominent colpocephaly, and in one patient even detected prenatally (P5). Comparison with imaging findings of previously reported patients (Table 1), confirmed the high prevalence of these findings (Table 2).
3.1.2 Molecular characterization
We identified a total of nine AP4B1 mutations in eight patients from seven families (Table 3, Figure 3, Supplementary Table S1). Two of these mutations (p.L222Cfs*31 and p.T387Rfs*30) have been previously reported (Abdollahpour et al., 2015; Abou Jamra et al., 2011; Bauer et al., 2012; Lamichhane, 2013; Tan et al., 2015; Tuysuz et al., 2014). In most patients (7/8), AP4B1 mutations were identified by exome sequencing. Four patients were of European descent, two patients were of mixed ethnicity, and two patients (siblings) were of South Asian (Tamil) origin. Consanguinity was reported in two families and shared ancestry in one (Table 1). Four patients (P1 and P4) had no history of shared ancestry and had compound heterozygous mutations (Tables 1 and 3). Interestingly, two unrelated individuals (P1 and P4) in our cohort shared the same compound heterozygous mutation (p.Arg406*/p.Leu443Pro).
| PATIENT | METHOD | MUTATION 1 | MUTATION 2 |
|---|---|---|---|
| P1 | Panel | c.1216C>T (p.Arg406*) | c.1328T>C (p.Leu443Pro) |
| P2 | ES | c.530_531insA (p.Asn178Glufs*20) | c.114-2A>C |
| P3 | ES | c.1345A>T (p.Arg449*) | c.1160_1161delCA (p.Thr387Argfs*30) |
| P4 | ES | c.1216C>T (p.Arg406*) | c.1328T>C (p.Leu443Pro) |
| P5 | ES | c.114-2A>G | c.114-2A>G |
| P6 | ES | c.304C>T (p.Arg102*) | c.304C>T (p.Arg102*) |
| P7 | ES | c.304C>T (p.Arg102*) | c.304C>T (p.Arg102*) |
| P8 | ES | c.664delC (p.Leu222Cysfs*31) | c.664delC (p.Leu222Cysfs*31) |
| Abdollahpour et al. (2015) (P1) | ES | c.1160_1161delCA (p.Thr387Argfs*30) | c.1160_1161delCA (p.Thr387Argfs*30) |
| Abdollahpour et al. (2015) (P2) | ES | c.1160_1161delCA (p.Thr387Argfs*30) | c.1160_1161delCA (p.Thr387Argfs*30) |
| Tuysuz et al. (2014) (P5) | Hmz/ES | c.664delC (p.Leu222Cysfs*31) | c.664delC (p.Leu222Cysfs*31) |
| Tuysuz et al. (2014) (P6) | Hmz/ES | c.664delC (p.Leu222Cysfs*31) | c.664delC (p.Leu222Cysfs*31) |
| Lamichhane (2013) | Panel | c.313delG (p.Ala105Argfs*52) | c.577G>A (p.Val193Ile) |
| Abou Jamra et al. (2011) (IV–2) | Hmz/sanger | c.487_488insTAT (p.Glu163Valfs*2) | c.487_488insTAT (p.Glu163Valfs*2) |
| Abou Jamra et al. (2011) (IV–4) | Hmz/sanger | c.487_488insTAT (p.Glu163Valfs*2) | c.487_488insTAT (p.Glu163Valfs*2) |
| Abou Jamra et al. (2011) (IV–5) | Hmz/sanger | c.487_488insTAT (p.Glu163Valfs*2) | c.487_488insTAT (p.Glu163Valfs*2) |
| Blumkin et al. 2011/Bauer et al. (2012) (P1) | Hmz/ES | c.664delC (p.Leu222Cysfs*31) | c.664delC (p.Leu222Cysfs*31) |
| Blumkin et al. 2011/Bauer et al. (2012) (P2) | Hmz/ES | c.664delC (p.Leu222Cysfs*31) | c.664delC (p.Leu222Cysfs*31) |
| Tan et al. (2015) (P1) | Panel | c.311_312delTGinsC (p.Leu104Profs*53) | c.577G>A (p.Val193Ile) |
| Helbig et al. (2016) | ES | c.617G>A (p.Arg206Gln) | c.617G>A (p.Arg206Gln) |
| Helbig et al. (2016) | ES | c.405_409delTGTCA (p.Tyr135*) | c.405_409delTGTCA (p.Tyr135*) |
| Karaca et al. (2015) | ES | c.955T>C (p.Phe319Leu) | c.955T>C (p.Phe319Leu) |
| Karaca et al. (2015) | ES | c.1282G>T (p.Glu428*) | c.1282G>T (p.Glu428*) |
- Nomenclature based on NM_001253852.1. The EXAC based minor allele frequencies for these variants range from 0 to 0.00018. ES (exome sequencing), Hmz (homozygosity mapping), Panel (multi-gene panel), Sanger (Sanger sequencing) See Supplemental Table S1 for complete genomic location, CADD scores, and minor allele frequencies.
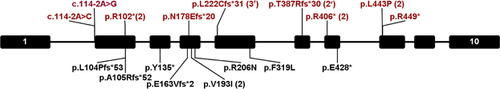
The literature includes two reports of patients with AP4B1 mutations in which little clinical information was provided (Helbig et al., 2016; Karaca et al., 2015). We included this data in our mutation summaries (Table 3, Figure 3, Supplementary Table S1), but did not in our summary of clinical characteristics (Table 1).
Including mutations identified in this report as well as the literature, there are now a total of 17 unique mutations in AP4B1, five of which are recurrent (Table 3). The majority (11/17) are frameshifting or nonsense mutations. Two splice site mutations are at the same splice acceptor site, and four missense mutations have been described. The most common mutation in AP4B1 is c.664delC. One mutation (p.Thr387Argfs*30) is seen in both homozygous (P1 in [Abdollahpour et al., 2015]) and compound heterozygous state (P3).
4 DISCUSSION
We report a series of eight new patients with bi-allelic AP4B1 mutations from mostly non-consanguineous families, and combine detailed clinical, imaging, and molecular characterization of these patients with the 11 patients previously reported. Together, these 19 patients begin to define a core set of features (Table 2).
These consist of a combination of early developmental delay and intellectual disability, prominently delayed motor and speech development, infantile-onset hypotonia that progresses to symmetric spastic diplegia with loss of independent ambulation, mild (−2 to −3 SD) microcephaly and episodes of stereotypic laughter. Epilepsy is a variable manifestation and is seen in about half of all patients. Although some of the individuals have overlapping facial features, a “recognizable” dysmorphic facial phenotype is not readily apparent (Figure 1).
Characteristic brain imaging findings include a triad of thin corpus callosum, delayed myelination or loss of supratentorial white matter, and associated ventriculomegaly (Figure 2).
Mutations in AP4B1 were detected by exome sequencing or multi-gene panels (Table 3). An additional four individuals were identified in large cohorts referred for exome sequencing for epilepsy (Helbig et al., 2016) or developmental brain malformations (Karaca et al., 2015) without detailed clinical data available. The majority (11/17) of mutations are frameshifting or nonsense mutations that likely lead to nonsense-mediated RNA degradation (Hirst et al., 2013). This strongly suggests the molecular mechanism behind this autosomal-recessive disease is loss of function. The c.664delC (p.Leu222Cysfs*31) mutation was found in five patients from three families, making this the most common mutation in AP4B1. This mutation has thus far only been seen in homozygous individuals with Arab/Turkish ancestry, so it may be specific to this population. It could also represent a mutational hotspot as this cytosine base is located within a short tract of thymine bases.
Two unrelated patients in our cohort shared identical mutations: c.1216C>T (p.Arg406*)/c.1328T>C (p.Leu443Pro). The ExAC minor allele frequency of c.1216C>T is 0.01153% (14 out of 121372 alleles) and c.1328T>C is 0.00083% (1 out of 121244 alleles), both of these variants are rare. We cannot explain this observation at the present, but speculate that only a limited number of missense mutations will be deleterious enough to AP-4 function to produce disease. Experiments reproducing this and other AP4B1 missense mutations in model systems are needed to address this question.
Several individuals in our cohort were diagnosed by exome sequencing within 3–4 months of each other. This suggests that this disorder is likely under-diagnosed at the present, and the increasing use of exome sequencing will undoubtedly lead to the identification of more individuals with this disorder. Due to overlapping clinical features, patients with AP4B1 related SPG47 may be misdiagnosed as having cerebral palsy. Indeed several individuals in our cohort were thought to have cerebral palsy prior to genetic diagnosis. Clues that can help distinguish AP4B1-related spastic paraplegia from cerebral palsy include the brain MR findings, the progressive nature of the disease and the severity of the speech delay. The early onset of developmental delay and microcephaly, when present, are also somewhat atypical for cerebral palsy. Due to the 25% recurrence risk for this autosomal-recessive disorder, it is a critically important diagnosis to make.
Seven out of the eight newly identified patients are female, and in the total cohort of 19 patients, only six are males. This cause of this sex imbalance is currently unknown. A skewed sex ratio in cerebral palsy has been previously noted, but in the opposite direction from what we observe. Cerebral palsy has been noted to occur 1.3–1.4 times more often in males than in females (Romeo et al., 2016). It is possible that males with biallelic AP4B1 mutations are more severely affected and do not survive long enough to be tested, although no difference in phenotypic severity is noted in the six males with AP4B1-related SPG47. It is also possible that males with the disease are less likely to receive testing because they are thought to have cerebral palsy.
Several of the families in our cohort identified each other using social media. In the current era of exome sequencing, social media has become an increasingly common and effective tool for families with rare disorders. The efforts of the families in this study, represented by the CureSPG47 foundation (www.CureSPG47.org) have significantly enabled this collaborative research effort.
There are striking similarities between the phenotypes that arise from mutations in AP4B1 and the other three subunits of AP-4. Patients with AP4E1 (Moreno-De-Luca et al., 2011), AP4M1 (Jameel et al., 2014; Tuysuz et al., 2014; Verkerk et al., 2009), and AP4S1 (Hardies et al., 2015; Tessa et al., 2016) mutations also present with spastic para- or tetraplegia, intellectual disability, delayed or absent speech development, microcephaly, seizures and febrile seizures in particular, and many have pseudobulbar signs such as stereotypic laughter, indicating a common phenotype that arises from loss of AP-4. Thus enhanced understanding of AP4B1-SPG47 will likely benefit the other AP-4 related disorders.
The AP-4 complex has been shown to be involved in protein trafficking from the trans-Golgi network to early and late endosomes but also the plasma membrane (Bonifacino, 2014; Hirst et al., 2013). In neuronal models, AP-4 was found to mediate AMPA receptor trafficking with Ap4b1−/− mice showing mislocalization of AMPA receptors to distal axons, where they were found within autophagosomes (Matsuda et al., 2008). The link to neuronal autophagy is compelling given the clinical overlap between AP4B1-SPG47 and the congenital disorders of autophagy (Ebrahimi-Fakhari et al., 2016), in which the long white matter tracts are often prominently affected. Further research is needed to understand the molecular mechanisms by which loss of AP-4 function leads to degeneration of the long white matter tracts and other manifestations.
In summary, AP4B1-associated HSP and other AP-4-deficiency syndromes should be suspected in infants and children with hypotonia progressing to spastic paraplegia, delayed motor and speech development, and suggestive findings on brain imaging particularly thinning of the posterior aspect of the corpus callosum. Given the rarity of many forms of HSP, patients with childhood-onset HSP may be mistakenly diagnosed with cerebral palsy, particularly if there is no family history or history of consanguinity that would suggest a genetic etiology.
ACKNOWLEDGMENTS
The authors thank the patients and families who participated in this study. The authors are grateful to the CureSPG47 organization (www.CureSPG47.org) for endorsing and supporting this study. The authors thank Lea Florentino (Boston Children's Hospital) and Ana Westenberger (University of Lübeck) for assistance. D.E.-F. received support from the Fred Lovejoy Research and Education Fund. W.K.C. received support from the Simons Foundation. J.T.B received support from the Burroughs Wellcome Fund Career Award for Medical Scientists and the Arnold Lee Smith Endowed Professorship for Research Faculty Development. Patient 7 and 8 were identified through the Deciphering Developmental Disorders (DDD) study. The following is acknowledged: This study makes use of data generated by the DECIPHER community. A full list of centers who contributed to the generation of the data is available from http://decipher.sanger.ac.uk and via email from [email protected] Funding for the project was provided by the Wellcome Trust. The DDD study presents independent research commissioned by the Health Innovation Challenge Fund (grant number HICF-1009-003), a parallel funding partnership between the Wellcome Trust and the Department of Health, and the Wellcome Trust Sanger Institute (grant number WT098051). The views expressed in this publication are those of the author(s) and not necessarily those of the Wellcome Trust or the Department of Health. The study has UK Research Ethics Committee approval (10/H0305/83, granted by the Cambridge South REC, and GEN/284/12 granted by the Republic of Ireland REC). The research team acknowledges the support of the National Institute for Health Research, through the Comprehensive Clinical Research Network.