Novel recessive PDZD7 biallelic mutations in two Chinese families with non-syndromic hearing loss
Abstract
Autosomal recessive non-syndromic hearing loss (ARNSHL) is a highly heterogeneous genetic condition. PDZD7 has emerged as a new genetic etiology of ARNSHL. Biallelic mutations in the PDZD7 gene have been reported in two German families, four Iranian families, and a Pakistani family with ARNSHL. The effect of PDZD7 on ARNSHL in other population has yet to be elucidated. Two Chinese ARNSHL families, each of which had two affected siblings, were included in this study. The families underwent target region capture and high-throughput sequencing to analyze the exonic, splice-site, and intronic sequences of 128 genes. Furthermore, 1751 normal Chinese individuals served as controls, and 122 Chinese families segregating with apparent ARNSHL, who had been previously excluded for variants in the common deafness genes GJB2 and SLC26A4, were subjected to screening for candidate mutations. We identified a novel homozygous missense mutation (p.Arg66Leu) and novel compound heterozygous frameshift mutations (p.Arg56fsTer24 and p.His403fsTer36) in Chinese families with ARNSHL. This is the first report to identify PDZD7 as an ARNSHL-associated gene in the Chinese population. Our finding could expand the pathogenic spectrum and strengthens the clinical diagnostic role of the PDZD7 gene in ARNSHL patients.
1 INTRODUCTION
Hearing loss is a prevalent sensorineural disorder that is characterized by clinical and genetic heterogeneity. It is estimated that 70% of the genetic forms of hearing loss are nonsyndromic and the remaining 30% are syndromic (Morton & Nance, 2006).Variations in many genes can cause either non-syndromic hearing loss or syndromic hearing loss. PDZD7 encodes a PDZ domain-containing scaffold protein, that was recently implicated as a modifier and candidate gene in human USH2 (Ebermann et al., 2010), and it is an autosomal recessive genetic hearing loss (ARNSHL)-associated gene (Booth et al., 2015; Le Quesne Stabej et al., 2017; Schneider et al., 2009; Vona et al., 2016). The PDZD7 gene was first identified as being responsible for ARNSHL in a consanguineous German family in 2009. A homozygous disruption of PDZD7 by reciprocal translocation was found in a 9-year-old boy with non-syndromic congenital sensorineural hearing loss (Schneider et al., 2009). Clinical tracking of this patient over 6 years showed that the proband demonstrated stable sensorineural hearing loss and healthy retinas at the age of 15.5 years (Vona et al., 2016). To date, only seven biallelic mutations of the monogenic PDZD7 have been validated—in two German families, four Iranian families, and a Pakistani family with ARNSHL (Booth et al., 2015; Le Quesne Stabej et al., 2017; Vona et al., 2016). Outside of the Iranian, German and Pakistani populations, little is known about the effect of PDZD7 on ARNSHL. More reports will be required in order to understand the complete phenotypic spectrum of the PDZD7 gene.
Here, we are the first to show a novel homozygous missense mutation and compound heterozygous frameshift mutations of the PDZD7 gene in two unrelated Chinese families with ARNSHL. Our work could expand the pathogenic spectrum and strengthen our understanding of the mechanism by which PDZD7 causes ARNSHL.
2 MATERIALS AND METHODS
2.1 Ethics statement
This study was approved by the Committee of Medical Ethics of Chinese People's Liberation Army (PLA) General Hospital. We obtained written informed consent from all the participants in this study. Written informed consent was obtained from the next of kin on the behalf of the minors/children who participants involved in this study.
2.2 Subjects
Two two-generation Chinese families (CN-0501754 and CN-1507352) with recessive non-syndromic hearing loss (NSHL) were introduced to the Department of Otolaryngology and Head and Neck Surgery at the Chinese People's Liberation Army (PLA) General Hospital. The subjects underwent a full medical history and comprehensive audiological evaluation, including otoscopic examination, pure-tone audiometry, tympanometry, acoustic reflex, distortion product evoked otoacoustic emissions (DPOAEs), and auditory brainstem responses (ABRs). Temporal bone Computed Tomography (CT) scans were also performed.
The two patients in family CN-1507352 underwent comprehensive ophthalmic evaluation, including visual acuity, direct funduscopy, visual field test, and electroretinogram (ERG) analysis. The proband (II–1) additionally underwent rotational chair testing (RCT) and colic vestibular evoked myogenic potential (cVEMP) testing.
Air conduction (AC) thresholds were bilaterally determined at octave frequencies of 0.25–8.0 kHz. The AC average thresholds at conversational frequencies of 0.5, 1, 2, and 4 kHz were measured and used to define the severity of the hearing loss. Hearing levels were labeled subtle (16–25 dB), mild (26–40 dB), moderate (41–70 dB), severe (71–95 dB), or profound (95 dB) (Kim et al., 2015).
2.3 Targeted gene capture and high throughput sequencing
Genomic DNAs of the probands, their brothers, and parents were extracted from peripheral blood samples using the Blood DNA kit (TIANGEN BIOTECH, Beijing, China).
Targeted gene capture and high throughput sequencing have been described in detail previously (Wang et al., 2014). In brief, we used a targeted genomic enrichment platform to simultaneously capture exons, splicing sites and immediate flanking intron sequences of 128 known deafness genes (Supplementary Table S1). The probands underwent targeted region sequencing, which built the DNA library. Universal primers for PCR amplification were adopted, after which sequencing of the PCR products was carried out on an Illumina HiSeq 2000 (San Diego, CA). The test platform examined >95% of the target gene with a sensitivity of >99%. Point mutations, micro-indels, and duplications (<20 bp) could be detected simultaneously. The data were not analyzed for copy number variants (CNVs).
2.4 Mutation analysis and control screening
Sanger sequencing was used to identify participants segregating causative variants in the candidate gene. PCR was performed with a PE9700 thermocyclers (Applied Biosystems, Foster City, CA). Sequence analysis was performed on an automated sequencer (ABI 3730, Applied Biosystems) for both affected and normal individuals. Nucleotide alterations including mutations and polymorphisms were identified by sequence alignment with the NCBI Reference Sequence (NM_001195263.1) using the DNAStar software versions 5.0 (DNASTAR, Madison, WI).
A total of 1,751 normal Chinese individuals served as controls, and 122 Chinese families segregating apparent ARNSHL, who had previously been shown not to have abnormalities in common deafness genes, were ascertained for screening candidate mutations.
2.5 In silico analysis and molecular structural modeling
In this study, we used SIFT and PolyPhen software to analyze possible changes in the protein function and polymorphism phenotyping. Clustal X1.8 software was used to compare the human wild-type PDZD7 protein sequence with the orthologs from Homo sapiens, Macaca fascicularis, Marmota marmot marmota, Macaca mulatta, Acinonyx jubatus, Pan paniscus, Gekko japonicus, and Mus musculus and to examine the evolutionary conservation and the structural predication for the protein. We identified feasible template structures for PDZD7 using SWISS-MODEL (http://www.swissmodel.expasy.org). The three-dimensional (3D) protein modeling for wild-type and mutant PDZD7 proteins were obtained using the Swiss-PdbViewer 4.1 molecular visualization software.
3 RESULTS
3.1 Clinical evaluation
Families CN-1507352 and CN-0501754 are both two-generation families segregating ARNSHL (Figure 1). The age of onset varied from 11 months to 5 years.
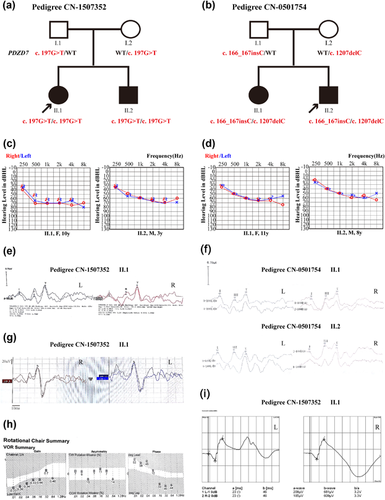
In family CN-1507352, the affected siblings (Figure 1a, II–1, 2) were a 10-year-old girl and a 3-year-old boy. They showed bilaterally symmetrical mild hearing loss at 250 Hz and moderate to severe hearing loss between 500 and 8000 Hz (Figure 1c). Individual II–1(10y) had worse hearing thresholds than II–2(3y). Temporal bone CT scans were normal in subject II–2. Their parents showed normal bilateral hearing (Supplementary Figure S1). After two years, audiological testing of the proband (II–1) at the age of 12 and her 5-year-old brother (II–2) showed symmetrical stable hearing loss. The subjects both exhibited normal latency and amplitude of ABR waves I, III, and V (Figure 1e; Supplementary Table S2). Funduscopic examination and visual field testing were normal (Supplementary Figure S2). ERG testing showed a normal response (Figure 1i). The proband had normal cVEMP amplitude and latencies (Figure 1g). The RCT demonstrated normal vestibular ocular reflex (Figure 1h). The vestibular function tests were not well-tolerated by the 5-year-old patient (II–2).
In family CN-0501754, the affected siblings (Figure 1b, II–1, 2) were an 11-year-old girl and an 8-year-old boy. They showed bilaterally symmetrical mild hearing loss at 250 Hz and moderate hearing loss between 500 and 8000 Hz. Individual II–1(11y) show slightly worse hearing than II–2(8y). They exhibited normal latency and amplitude of ABR waves I, III, and V (Figures 1d and 1f). Temporal bone CT scans were normal in subject II–1. A telephone follow-up call was immediately conducted after we identified the disease-causing gene. The affected members (Figure 1b, II–1, 2) are now a 22-year-old woman and a 19-year-old man. Both adult patients reported that they had no symptoms of night blindness or a reduction in their visual field. They also reported no vestibular disorders and no progression in hearing loss during the 11-year follow up period.
3.2 Targeted high-throughput sequencing
Approximately 619 kb of exons and adjacent intronic regions of the deafness panel were captured and sequenced using next generation sequencing in the probands. The average depth of cover for targeted regions exceeded 190×, with >97% of based having >30× depth of coverage.
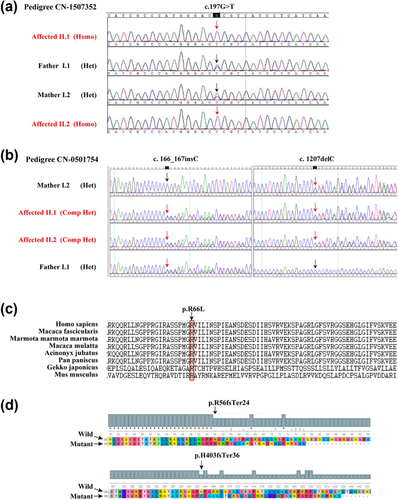
For the proband of family CN-1507352, a total of 253 variants were identified, 107 of which were nonsynonymous variants, splice acceptor and donor site mutations, and coding indels that were more likely to be functional mutations; only six of the 107 coding variants were an allele frequency of less than 0.01 in the 1000 human genome and a local dataset. A homozygous variant leading to amino acid change were detected in PDZD7 c.197G>T (p.R66L, Arg66-to-Leu) (Supplementary Tables S3 and S4). We further performed in silico analyses, and a missense variant, c.197G>T in exon 2 of PDZD7 was predicted to be “Deleterious” by SIFT, “Probably damaging” by PolyPhen, “Conserved” by GERP++ and Phylop software (Table 1). These results indicated that this novel mutation could be the cause of the hearing loss in this Chinese family. Sanger sequencing revealed that the mutation segregates with the phenotype, and unaffected parents were heterozygous carriers (Figures 1a and 2a).
| AF in database | AF in our study | Prediction information | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Family no. | Gene | RefSeq | Nucleotide | Amino acid | Zygosity | ExAC | gnomAD | ARNSHL | Control | SIFT (score) | Polyphen (score) | Mutation taster | GERP ++ (score) | PhyloP (score) |
| CN-1507352 | PDZD7 | NM_001195263.1 | c.197G>T | p.Arg66Leu | Hom | 0 | 0 | 0/244 | 0/3502 | Deleterious (0) | Probably damaging (0.994) | Disease causing | Conserved (5) | Conserved (2.317) |
| CN-0501754 | PDZD7 | NM_001195263.1 | c.1207delC | p.His403Ilefs*36 | Comp Het | 0 | 0 | 0/244 | 0/3502 | N/A | N/A | N/A | N/A | N/A |
| c.166_167insC | p.Arg56Profs*24 | 0 | 0.0000543 | 0/244 | 0/3502 | |||||||||
- N/A, not available; AF, allele frequency of existing variant; ARNSHL, autosomal recessive non-syndromic hearing loss; Hom, homozygous mutation; Comp Het, compound heterozygous mutation.
Subsequently, for the proband of another family (CN-0501754), a total of 298 variants were identified, 176 of which were nonsynonymous variants, splice acceptor and donor site mutations, and coding indels that were more likely to be pathogenic mutations; only 8of the 176 coding variants were an allele frequency of less than 0.01 in the 1000 human genome and local dataset. Two heterozygous variants leading to amino acid change were detected in PDZD7 c.166_167insC (p.Arg56Profs*24) (rs760796701) and c.1207delC (p. His403Ilefs*36) (Supplementary Tables S2–S4). The c.166_167insC was found in heterozygous form in 15/276238 individuals in the gnomAD browser. The other variant was not available. The PDZD7 mutation p.Arg56Profs*24 in heterozygous state was reported previously as a retinal phenotype modifier in Usher syndrome. Compound heterozygous c.166_167insC and c.1207delC mutations of PDZD7 were novel compound heterozygous frameshift mutations. These mutations were validated by Sanger sequencing and were found to co-segregate with the phenotype of all family members (Figures 1b, 2b, and 2d).
Furthermore, we screened 1,751 normal Chinese individuals and 122 unrelated Chinese families segregating as apparent ARNSHL but failed to detect the three confirmatory mutations. The c.197G>T (p.R66L) occurs at highly conserved residues (Figure 2c, Table 1).
3.3 Molecular structural modeling of the R66L amino acid substitution
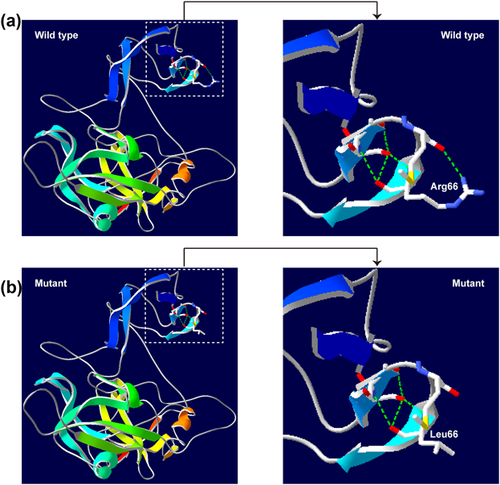
A 3D structural model of PDZD7 was predicted with SWISS-MODEL to determine whether the novel p.R66L missense mutation in an ARNSHL Chinese family affects the structure of PDZD7. The sequence identity between the target and template was 26.59%. The R66L molecular model covered the target sequence of PDZD7 (residues 4–294). Using Swiss-Pdb Viewer 4.1, the mutation was predicted to result in the loss of an H-bond between Arg 66 and Met 64 due to the substitution of arginine to leucine at position 66, possibly perturbing the amino acid side chain (Figure 3).
4 DISCUSSION
PDZD7 encodes a PDZ domain-containing scaffold protein; the largest transcript (NM_001195263.1) of PDZD7 encodes 17 exons, and it has been implicated as an ARNSHL-associated gene (Booth et al., 2015; Le Quesne Stabej et al., 2017; Schneider et al., 2009; Vona et al., 2016) and a modifier and candidate gene for human USH2 (Ebermann et al., 2010). In this study, next generation sequencing (NGS) was used to find the disease-causing gene of two Chinese families with ARNSHL (Figure 1a and 1b), and we identified two biallelic mutations in the PDZD7 gene. Furthermore, we determined that the both biallelic mutations are causative. This prediction was supported by the fact that the mutation co-segregated with the disease status. PDZD7 mutations are rare causes of recessive deafness, and only seven biallelic mutations have been detected, as shown in Table 2. Of our two families, one showed a novel homozygous missense mutation (c.197G>T, p.Arg66Leu), and the other a compound heterozygous frameshift mutation (c.166_167insC, p.Arg56Profs*24, and c.1207delC, p. His403Ilefs*36). PDZD7 is expressed in the stereocilia ankle-link region of the mechanosensitive structure. The PDZD7 protein localization at the connecting cilium region has been shown to play a role in inner ear hair cell development (Ebermann et al., 2010). In mice, the abnormal expression of the Pdzd7gene causes disorganization of hair bundles and may affect the establishment of mechanotransduction responses, which likely results in hearing loss (Zou et al., 2014). PDZD7 is a three-PDZ domain protein with a central proline-rich region. Multi-PDZ domain-containing scaffolds could play a complementary role in the structure of the ankle-links for the mutant type (Grati et al., 2012). The three mutations of our study lie outside functional domains, but the c.197G>T(p.R66L) mutation is conserved and predicted to have a deleterious result in the loss of an H-bond between Arg 66 and Met 64 due to the substitution of arginine to leucine at position 66, possibly perturbing the amino acid side chain (Figure 2), and the two frameshift mutations lead to a truncated protein and could affect the PDZ2 and PDZ3 domains.
| Origin | Mutation type | Genotype | HGVS.c | HGVS.p | Exon | Age of the indexes | Hearing phenotype | Reference |
|---|---|---|---|---|---|---|---|---|
| China | Missense | Homo | c.197G>T | p.R66L | Exon 2 | 10 | Down sloping/ moderate to severe | Present study |
| Pakistani | Frameshift | Homo | c.226 + 2_266 + 5delTAGG | ? | Intron 2 | N/A | Moderate | Le Quesne Stabej et al. (2017) |
| Iranian | Missense | Homo | c.307G>C | p.G103R | Exon 3 | 36 | Down sloping/ moderate to severe | Booth et al. (2015) |
| Iranian | Missense | Homo | c.682G>A | p.G228R | Exon 5 | 33 | Flat/ moderate to severe | Booth et al. (2015) |
| Iranian | Missense | Comp Het | c.854T>G | p.M285R | Exon 6 | 38 | Down sloping/moderate to severe | Booth et al. (2015) |
| Nonsense | c.1500C>A | p.T500X | Exon 9 | |||||
| China | Frameshift | Comp Het | c.1207delC | p.H403 fs | Exon 8 | 11 | Down sloping/moderate | Present study |
| Frameshift | c.166_167insC* | p.R56 fs | Exon 2 | |||||
| Iranian | Nonsense | Homo | c.1576C>T | p.Q526X | Exon 10 | 23 | Down sloping/severe to profound | Booth et al. (2015) |
| German | Reciprocal translocation | Homo | t(10;11) | ? | Intron 10 | 15 | Down sloping/ moderate to severe | Schneider et al. (2009) |
| Vona et al. (2016) | ||||||||
| German | Nonsense | Comp Het | c. 1648C>T | p.Q550X | Exon 10 | 10 | Down sloping/mild to severe | Vona et al. (2016) |
| Frameshift | c.2107del | p.S703 fs | Exon 14 |
- * Ebermann et al. (2010) reported the PDZD7 monoallelic frame-shift mutation with a homozygous USH2A mutation in one of two USH2 affected sisters, who had earlier onset and more severe retinitis pigmentosa.
- N/A, not available.
Mutations in this gene have been documented to cause moderate-severe gently downward sloping hearing loss. Our affected siblings also showed bilaterally symmetrical moderate-severe gently down sloping hearing loss. In addition, our results showed that individual II–1(10y) of family CN-1507352 had slightly worse hearing thresholds than individual II–2(3y). Furthermore, individual II–1(11y) of family CN-0501754 was also worse than II–2(8y), especially in the low frequencies. This may suggest that PDZD7 mutations are associated with milder progressive hearing loss.
In addition, PDZD7 is a member of the Usher quaternary protein complex (PDZD7, USH2A, GPR98 and WHRN), and localization in the ankle-link makes it a good candidate for Usher syndrome type 2 (USH2) (Chen, Zou, Shen, Zhang, & Yang, 2014; Zou et al., 2014). USH2 is the most common Usher syndrome type. USH2 patients present with congenital moderate to severe hearing loss, retinitis pigmentosa (RP), and normal vestibular function. RP usually develops from the second decade onwards (Besnard et al., 2012; Chen et al., 2014). Early symptoms of RP include night blindness and loss of peripheral vision. However, to date, only monoallelic mutations in the PDZD7 gene act as a retinal disease modifier of homozygous USH2A mutations and a contributor of digenic inheritance with GPR98 in USH2 patients (Ebermann et al., 2010). The PDZD7 mutation p.Arg56Profs*24 (c.166_167 insC) was suggested to be a retinal phenotype modifier in Usher syndrome in its heterozygous state. No biallelic mutations of PDZD7 were found in USH2 (Aparisi et al., 2014; Besnard et al., 2012). In this study, we did not find other functional candidate gene variants of classical Usher syndrome genes by targeted region capture and high throughput sequencing. At present, the oldest patient reported with ARNSHL-associated PDZD7 biallelic mutations is 38 years old (Table 2), and this patient showed no abnormalities in funduscopy (Booth et al., 2015). In our study, the age of the ARNSHL affected individuals was younger than 11 years of age during examinations. Furthermore, after 11 years, the two patients of family CN-0501754 were 22 and 19 years of age. At present, they describe no signs of visual or vestibular disorder and no progression in hearing loss from 2005 to now. PDZD7 is essential for the normal localization of USH2A, GPR98, and WHRN in inner ear hair cells. The disruption of PDZD7 expression affects the integrity of the USH2 complex (Chen et al., 2014). The hair cells of Pdzd7−/− mice display bundle morphological defects and abnormal mechanotransduction (MET) processes in OHCs while vestibular hair cells have normal MET responses. Furthermore, in reported Pdzd7 mutant mice, PDZD7 plays a limited role in localizing USH2 proteins in photoreceptors (Zou et al., 2014). Overall, our data support the contribution of PDZD7 biallelic mutations to the etiology of ARNSHL in humans.
This is the first study to identify PDZD7 as an ARNSHL-associated gene in the Chinese population. The study is significant because it leads to a better understanding of PDZD7 biallelic mutations on phenotypic outcome and strengthens the clinical diagnostic role of the PDZD7 gene in ARNSHL patients.
ACKNOWLEDGMENTS
We thank the families for their invaluable cooperation and participation. We thank all colleagues in the ENT departments who provided the patient samples. We thank Cui Zhao for the DNA extraction and Sanger sequencing. We thank the families for their participation. This work was supported by grants from the National Key Basic Research Program of China (No. 2014CB943001), the National Natural Science Foundation of China (No. 81120108009 and 81530032), and the China Postdoctoral Science Foundation (No. 2015M572766).
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.
AUTHORS’ CONTRIBUTIONS
QW JG: conceived and designed the experiments. JG, HW, and LL: performed the experiments. JG and HW: analyzed the data. LL, LW, JY, LX, ZY, and WX: contributed reagents/materials/analysis tools. JG and QW: wrote the paper. JG, LZ, DW, and QW: critically read and discussed the manuscript.