Identification of a de novo variant in CHUK in a patient with an EEC/AEC syndrome-like phenotype and hypogammaglobulinemia
Abstract
The cardinal features of Ectrodactyly, Ectodermal dysplasia, Cleft lip/palate (EEC), and Ankyloblepharon-Ectodermal defects-Cleft lip/palate (AEC) syndromes are ectodermal dysplasia (ED), orofacial clefting, and limb anomalies. EEC and AEC are caused by heterozygous mutations in the transcription factor p63 encoded by TP63. Here, we report a patient with an EEC/AEC syndrome-like phenotype, including ankyloblepharon, ED, cleft palate, ectrodactyly, syndactyly, additional hypogammaglobulinemia, and growth delay. Neither pathogenic mutations in TP63 nor CNVs at the TP63 locus were identified. Exome sequencing revealed de novo heterozygous variants in CHUK (conserved helix-loop-helix ubiquitous kinase), PTGER4, and IFIT2. While the variant in PTGER4 might contribute to the immunodeficiency and growth delay, the variant in CHUK appeared to be most relevant for the EEC/AEC-like phenotype. CHUK is a direct target gene of p63 and encodes a component of the IKK complex that plays a key role in NF-κB pathway activation. The identified CHUK variant (g.101980394T>C; c.425A>G; p.His142Arg) is located in the kinase domain which is responsible for the phosphorylation activity of the protein. The variant may affect CHUK function and thus contribute to the disease phenotype in three ways: (1) the variant exhibits a dominant negative effect and results in an inactive IKK complex that affects the canonical NF-κB pathway; (2) it affects the feedback loop of the canonical and non-canonical NF-κB pathways that are CHUK kinase activity-dependent; and (3) it disrupts NF-κB independent epidermal development that is often p63-dependent. Therefore, we propose that the heterozygous CHUK variant is highly likely to be causative to the EEC/AEC-like and additional hypogammaglobulinemia phenotypes in the patient presented here.
1 INTRODUCTION
Ectrodactyly ectodermal dysplasia-cleft lip/palate (EEC) syndrome (OMIM 604292) is a rare genetic ectodermal dysplasia (ED) syndrome. Patients with EEC syndrome present with variable features of ectrodactyly, defects in skin, hair, and nails, and orofacial clefts (OFC). A related ED syndrome, Ankyloblepharon-ectodermal defects-cleft lip/palate (AEC) syndrome (OMIM 106260), is characterized by ankyloblepharon (fusion of the eyelids), severe malformations of ectoderm-derived tissues, such as skin, hair, nails, sweat glands, OFC, and limb anomalies (Rinne, Brunner, & van Bokhoven, 2007; Sutton et al., 2009). The clinical findings of EEC and AEC can overlap. For instance, ankyloblepharon, which is pathognomonic for AEC syndrome has been reported in some patients who were classified as EEC syndrome (Berdón-Zapata et al., 2004). In contrast to EEC syndrome, severe limb malformations such as ectrodactyly were not considered to be typical for the AEC syndrome phenotype. However, syndactyly and hammertoe deformities appeared to be surprisingly common (17/18 cases) in a cohort of AEC patients, suggesting that mild limb anomalies are a significant factor in AEC syndrome (Sutton et al., 2009). Hearing loss which has been reported in more than 90% of the AEC patients (Cole et al., 2009) rarely occurs in EEC patients also (Rinne, Hamel, van Bokhoven, & Brunner, 2006). ED symptoms such as skin erosions, sparse hair, and nail dysplasia are more severe in AEC than in EEC. Many AEC and EEC patients do not present with any OFC features. Patients with overlapping features of both EEC and AEC have been described (Rinne et al., 2006). For instance, Celik et al. (2011) reported a neonate with diffuse skin erosions, which are suggestive of AEC, while no ankyloblepharon or OFC, but with ectrodactyly and polydactyly, which are severe limb abnormalities and suggestive of EEC. Clinical heterogeneity is typical for both syndromes (Rinne et al., 2006).
Mutations in the transcription factor p63 encoded by TP63 are the underlying cause in the majority of diagnosed EEC and AEC syndrome cases. The p63 protein is important for the normal development of ectoderm-derived tissues. It plays an essential role in the development of the epidermis, the limbs, urinary system, and orofacial structures. TP63 is located on chromosome 3q28 (Celli et al., 1999; Yang et al., 1998) and encodes several p63 isoforms resulting from the use of two different promoters (TA and ΔN) and due to extensive alternative splicing routes. All the isoforms contain a DNA-binding domain (DBD) and an Oligomerization domain (OD), but they differ in their transactivation domain, the C-terminal sterile alpha-motif (SAM), and auto-regulating transactivation inhibiting domain (TID). Striking genotype-phenotype correlations have been observed for TP63 mutations in EEC and AEC syndromes. EEC syndrome mutations are located in the DBD of p63, which disrupts the p63 binding to DNA and hence, affect transactivation (Celli et al., 1999); while mutations leading to AEC are mainly seen in the SAM and TID domain as well as in the N-terminus that is unique for ΔN-isoforms of p63 (McGrath et al., 2001; Rinne et al., 2008).
Phenotypically related disorders are often caused by functionally related genes (Oti, Huynen, & Brunner, 2008). TP63 and its target genes constitute a gene network governing ectodermal development. Mutations in these p63 target genes and regulatory elements are known to give rise to various conditions that have overlapping features with the p63 associated syndromes. For example, IRF6 is a direct transcriptional target of p63 (Thomason et al., 2010). Mutations in IRF6 are known to cause Van der Woude syndrome (VWS, OMIM #119300) and Popliteal Pterygium syndrome (PPS, OMIM #119500), which often present with cleft lip and/or cleft palate (CL/P) (Kondo et al., 2002). Similarly, mutations and genomic alterations of DLX5/6 genes that are controlled by p63 lead to split hand/foot malformations (Gurrieri & Everman, 2013; Kouwenhoven et al., 2010; Shamseldin, Faden, Alashram, & Alkuraya, 2012; Velinov et al., 2012).
The patient reported in the present study exhibited an EEC/AEC syndrome-like phenotype with additional hypogammaglobulinemia and growth delay, which are not typically included in p63 mutant phenotypes. Using trio exome sequencing, we identified three de novo variants, affecting CHUK (conserved helix-loop-helix ubiquitous kinase), PTGER4, and IFIT2. As CHUK is known to be regulated by p63 (Candi et al., 2006) and plays a role in epithelial development (Hu et al., 2001; Sil, Maeda, Sano, Roop, & Karin, 2004), we propose that the de novo mutation in CHUK is responsible for the ectodermal phenotype in this patient.
2 CLINICAL REPORT
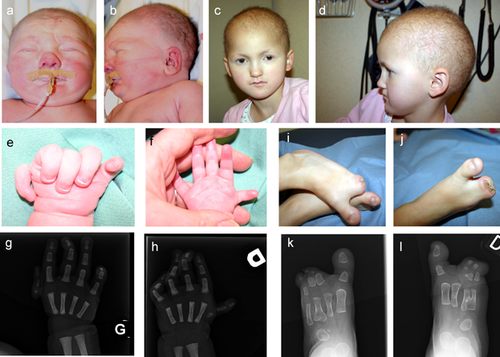
The patient, a 10-year-old girl, is the third child of healthy, non-consanguineous parents of Caucasian origin. The family history is unremarkable. The parents had no features of ED or hypogammaglobulinemia. At conception, the mother was 33 years old and the father 36 years old. During pregnancy, hypoplastic thumbs with reduced mobility and 3–5 syndactyly in feet with the absence of the second rays were observed. The child was born at 38.5 weeks. The growth parameters at birth were: weight 3,305 g, length 49 cm, and OFC 33.5 cm. The clinical examination revealed sparse hair, absent eyebrows and eyelashes, ankyloblepharon, buccal synechia, posterior cleft palate, retrognathia, and hypoplastic external genitalia. The limb anomalies seen prenatally were confirmed, and the nails were dysplastic. The X-rays showed complex hand and foot anomalies (Figure 1). The phenotype suggested EEC/AEC syndrome. The evolution of the child was marked by severe gastro-oesophageal reflux with swallowing problems, growth retardation, and frequent upper and lower respiratory tract infections. She was tube fed from birth until 4 months of age when a gastrostomy was required. At the age of 5 months, gammaglobulin deficiency and increased AST, ALT, and γ-GT were observed. The work-up showed normal cellular immunity and normal phagocytosis and discrete inflammation on hepatic biopsy. Since then, she has been under intravenous immunoglobulin therapy every 3 weeks and ursodeoxycholic acid. She developed bronchiectasis in the lower and middle right lobes and atelectasis of the left lower lobe. At the age of 7 years, coeliac disease was diagnosed. At the age of 10 years, her height was 118.8 cm (−3.5 SD), and the weight 20.5 kg (−3 SD). The deciduous teeth were conical and fragile. The panoramic radiograph showed agenesis of many permanent teeth.
3 METHODS
The coding region of the TP63 gene and the intron-exon boundaries were sequenced, and no pathogenic mutation was detected. Molecular karyotyping was performed using 250 K NspI Affymetrix SNP-array according to the manufacturer's instructions. No genomic rearrangement was observed.
3.1 Exome sequencing
Exome sequencing analysis was performed in a trio approach (patient and both parents), essentially as described before (Neveling et al., 2013). Exome capture was performed with the Agilent SureSelect Human All Exon v4 enrichment kit (Agilent Technologies, Santa Clara, CA). Whole-exome sequencing was performed on the Illumina HiSeq platform (BGI, Copenhagen, Denmark). Data were analyzed with BWA (read alignment) (Li & Durbin, 2009) and GATK (variant calling) (McKenna et al., 2010) software packages. Variants were annotated using an in-house developed pipeline. Prioritization of variants was done by an in-house designed “variant interface” and manual curation. Potentially causative variants were confirmed by Sanger sequencing.
3.2 Sanger sequencing
All 21 exons of CHUK were amplified in 11 additional patients with features reminiscent of EEC and AEC without mutations in TP63. Specific primers were designed through Primer3 (Untergasser et al., 2012) and sequences are shown in Supplementary Table SI. The PCR products were purified to remove excess primers and dNTPs using two hydrolytic enzymes, Exonuclease I (Exo I), and Thermosensitive Alkaline Phosphatase (FastAP) and sequenced.
3.3 Molecular structural modeling
The molecular effects of the IFIT2 mutation were studied using the experimentally solved protein structure (PDB file 4G1T) (Yang et al., 2012). For both CHUK and PTGER4, no known protein structure is available and therefore a homology model was created to study the CHUK mutation. The modeling script with standard parameters in the YASARA (Krieger, Koraimann, & Vriend, 2002) & WHAT IF Twinset (Vriend, 1990) was used to create this model based on PDB-file 4KIK (Liu et al., 2013). This template PDB-file contains the structure of human IKKβ, which has 56% sequence identity with CHUK over 593 residues. No modeling template for PTGER4 could be found. The mutation in this protein was analyzed using standard amino acid properties and knowledge obtained from the Uniprot database (UniProt Consortium, 2015; entry: P35408).
4 RESULTS
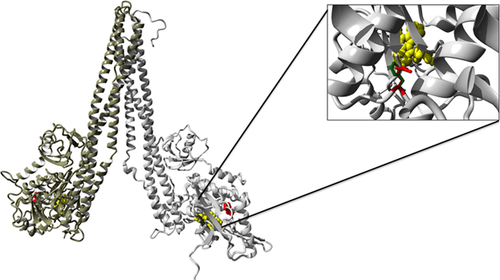
Given the EEC/AEC syndrome-like phenotype observed in this patient, we hypothesized that the p63 pathway is likely to be involved in the pathogenesis. Therefore, we first sequenced TP63 in the index patient, but no pathogenic mutation was detected. A genome-wide copy number analysis did not reveal any pathogenic CNVs. Subsequently, exome sequencing was performed in the index patient and the unaffected parents. Three heterozygous de novo variants were identified after the filtering steps as mentioned in Methods. These variants were present in PTGER4 (g.40691969C>T; p.Ser319Phe), IFIT2 genes (g.91066224A>G; p.Asn171Asp), and CHUK (g.101980394T>C; c.425A>G; p.His142Arg) (Table 1). All three variants were confirmed to be present in the patient and absent in both parents. The PTGER4 (p.Ser319Phe) variant occurs at a highly conserved nucleotide (phyloP: 5.69) and affects a moderately conserved amino acid. There is a large physicochemical difference between the reference amino acid serine and the variant amino acid phenylalanine (Grantham score: 155). The variant in IFIT2, p.Asn171Asp occurs at a weakly conserved nucleotide (phyloP: 0.29) and affects a moderately conserved amino acid. There is a small physicochemical difference between the reference amino acid asparagine and the variant amino acid aspartic acid (Grantham score: 23). The C-terminal part of IFIT2 folds into a super-helical structure containing nine tetratricopeptide repeats (TPR). The identified IFIT2 variant affects an amino acid in the TPR3 domain. Molecular structural modeling predicted the mutation to be on the surface of the protein, and it is unlikely to have a major effect on its function. The CHUK variant (p.His142Arg) occurs at a highly conserved nucleotide (phyloP: 4.32) and affects a highly conserved amino acid, close to the amino acid Asp144 responsible for the kinase activity (Figure 2). Using human IKKβ that shows 56% amino acid sequence identity with CHUK as a template, molecular structural modeling predicted that the affected amino acid is located at the core of the protein 3D structure, and the change is likely to affect the 3D structure of the protein (Figure2). In ExAC database, based on the observed and expected variants counts, the probability of intolerance of loss-of-function variations (pLI) was determined (Lek et al., 2016). Based on this, PTGER4 and CHUK are highly intolerant to loss-of-function mutations (pLI = 0.90 and 1.00, respectively), while IFIT2 is not intolerant at all (pLI = 0.0).
| Gene | Nucleotide positiona | Variant cDNA | Amino acid change | phyloP | CADD PHRED-like score | Probability of intolerance of LoF variations (pLI) |
|---|---|---|---|---|---|---|
| PTGER4 (NM_000958; P35408) | g.40691969C>T | c.956C>T | Ser319Phe | 5.693 | 18.57 | 0.90 |
| IFIT2 (NM_001547; O15111) | g.91066224A>G | c.511A>G | Asn171Asp | 0.36 | 13.1 | 0.00 |
| CHUK (NM_001278; P09913) | g.101980394T>C | c.425A>G | His142Arg | 4.382 | 23.6 | 1.00 |
- a Human genome build hg19 is used to annotate the variants.
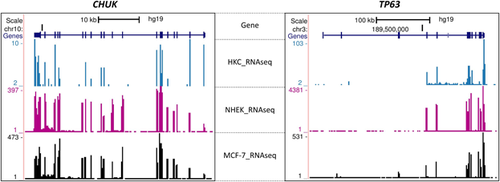
CHUK is known to be directly regulated by p63. To assess whether CHUK and TP63 share similar expression pattern in epithelial cells, we examined expression of these two genes using our previously reported RNA-Seq data of primary human keratinocytes (HKC) (Kouwenhoven et al., 2010) and publically available RNA-Seq data of normal human epidermal keratinocytes (NHEK) and those of a breast adenocarcinoma cell line MCF-7 (Dong et al., 2012; The ENCODE Project Consortium, 2012). As expected, both CHUK and TP63 were found to be expressed in all three cell lines (Figure 3). Mutations in CHUK have been associated with cocoon syndrome (OMIM #613630) and Bartsocas–Papas Syndrome (BPS, OMIM # 263650), two disorders that have overlapping but much severe phenotypes, as compared to EEC/AEC syndrome. Therefore, the variant in CHUK was considered to be most likely causative for the ED features. Eleven other patients with EEC or AEC who do not carry any TP63 mutation were assessed for mutations in CHUK. No pathogenic variant in CHUK was identified in these patients.
5 DISCUSSION
The index patient exhibited a phenotype similar to EEC/AEC syndrome with additional immunodeficiency. Thus, the TP63 gene which has been implicated in EEC and AEC syndrome was tested. No mutation in TP63 or CNV containing TP63 was identified. Subsequently, trio exome sequencing was performed and three de novo variants were identified in CHUK, PTGER4, and IFIT2 genes.
CHUK, also known as IKKα (Inhibitor of nuclear factor kappa-B kinase subunit alpha), encodes a serine/threonine kinase that plays a role in the NF-κB signaling pathway (Karin, DiDonato, Hayakawa, Rothwarf, & Zandi, 1997; Régnier et al., 1997). CHUK forms the IKK complex together with IKKβ and IKKγ (also known as NEMO). The canonical NF-κB pathway is dependent on the kinase activity of the IKK complex to phosphorylate and degrade inhibitors to activate NF-κB (Smahi et al., 2002; Yamamoto, Verma, Prajapati, Kwak, & Gaynor, 2003). It has been shown that both CHUK and IKKβ work in homodimer or heterodimer form (Huynh et al., 2000). The kinase domain of CHUK is located at the N-terminal region of the protein and is followed by a leucine zipper domain which facilitates homo- and hetero-dimerization. At the C-terminal region of CHUK, there is a NEMO (NF-κB essential modulator; IKKγ) binding region. The variant in CHUK (g.101980394T>C; c.425A>G; p.His142Arg) occurs in the core of the kinase domain, close to the amino acid Asp144 that is responsible for phosphorylation activity. CHUK is extremely intolerant to the loss-of-function variations. Therefore, this variant is highly likely to disrupt the kinase activity and be possibly causal.
Homozygous CHUK mutations that lead to severe ectodermal defects have been reported. A homozygous nonsense mutation of CHUK (c.1264C>T; p.Gln422X) was identified in two fetuses with severe fetal malformations in a Finnish family, named as cocoon syndrome (Lahtela et al., 2010). The mutation was inherited from heterozygous carrier parents, who were phenotypically unremarkable. Both the fetuses had an abnormal cyst in the cranial region, a large defect of the craniofacial area, and an omphalocele, as well as immotile and hypoplastic limbs. This CHUK mutation is predicted to result in a truncation after the kinase domain. Another homozygous CHUK mutation was identified in a patient with Bartsocas–Papas syndrome (BPS), which is considered to be a milder form of cocoon syndrome (Leslie et al., 2015). This patient presented multiple abnormalities including skin, limb, orofacial, and genital defects. This mutation is at the exon 10 splice acceptor site (c.934-2A>G) and also predicted to result in protein truncation after the kinase domain. In both cases, the described mutations are homozygous and lead to truncation of the dimer forming leucine zipper and very likely the complete loss of the protein. Heterozygous carriers of these loss-of-function alleles do not present any features of ED. In contrast, the CHUK heterozygous missense variant identified in the EEC/AEC-like patient with immunodeficiency in the current study appears to act in a dominant fashion. This variant is located in the kinase domain of CHUK, and is predicted to impair CHUK kinase activity, but not its capacity to engage in homomeric and heteromeric complexes. Therefore, it is likely that the mutant CHUK protein with this variant can heterodimerize with IKKβ in the IKK complex and exhibit a dominant negative effect toward the wild-type protein to affect the canonical NF-κB activation.
In agreement with this dominant-negative model of de-regulating the canonical NF-κB pathway, many mutations in NEMO including nonsense, frameshift, and sometimes missense mutations have been found to lead to ED syndromes with immune deficiency. These conditions include Incontinentia pigmenti (IP), Anhidrotic ED with immunodeficiency (EDA-ID), Anhidrotic ED with immune deficiency, osteopetrosis and lymphoedema (OL-EDA-ID), and hypomorphic mutations to a less severe phenotype of hypohidrotic/anhidrotic ectodermal dysplasia (HED/EDA) (Senegas, Gautheron, Maurin, & Courtois, 2015). Sometimes point mutations in NEMO can also give rise to immunodeficiency without ED phenotype. Furthermore, a frameshift mutation has also been reported in IKKβ to cause a severe combined immunodeficiency (SCID), that is characterized by recurrent infections and hypogammaglobulinemia/agammaglobulinemia (Pannicke et al., 2013). Mutations in other genes involved in NF-κB, namely EDA1, EDAR, and EDARADD have also been found to cause EDA (Smahi et al., 2002). These studies suggest that gene mutations perturbing the canonical NF-κB pathway can affect immunity. On the other hand, immunodeficiency observed in the patient in the current study was not reported in previous studies on cocoon syndrome and BPS. This raises the question whether the severe cocoon syndrome and BPS phenotypes prohibited the observation of possible immune phenotypes, or whether the complete loss of CHUK does not significantly affect the canonical NF-κB pathway. Although, the IKK complex and the NF-κB pathway are well known for their roles in immunohomeostasis, CHUK itself has a minor contribution to the overall kinase activity of the complex among the three IKK complex members (Descargues et al., 2008; Hu et al., 1999). CHUK-deficient mice show terminal differentiation problems of the epidermis, whereas the activation of NF-κB was not affected (Descargues et al., 2008; Li et al., 1999).
In addition to the role in the canonical NF-κB pathway, CHUK serves a key role in negative feedback of canonical and non-canonical NF-κB signaling (Razani et al., 2010; Shembade, Pujari, Harhaj, Abbott, & Harhaj, 2011). The negative feedback depends on the kinase activity of CHUK, but not on IKKβ nor NEMO (Razani et al., 2010). Both canonical and non-canonical NF-κB signaling, and the feedback loop seem to be involved in epidermal development (Smahi et al., 2002). The mutation identified in this study may affect the negative feedback of both canonical and non-canonical NF-κB signaling pathways.
Furthermore, CHUK directly regulates epidermal differentiation through an NF-κB independent route. It has been shown that a Nuclear Localization Signal (NLS) that is located in the kinase domain of CHUK is essential for CHUK to enter the nucleus and to directly regulate epidermal genes (Sil et al., 2004). This variant in the kinase domain may also disrupt the NLS that lies nearby, and abolish the ability of CHUK to enter the nucleus. This can lead to the complete loss of the NF-κB independent function of CHUK through which it regulates direct target genes important for epidermal differentiation.
Consistent with NF-κB independent role of CHUK in epidermal differentiation and development, CHUK is a direct p63 transcriptional target (Candi et al., 2006). Loss of Chuk in mice leads to multiple morphogenetic defects in limb and skeletal patterning, and in proliferation and differentiation of epidermal keratinocytes (Hu et al., 1999; Takeda et al., 1999), which is very similar to phenotypic features observed in Tp63-knockout mice (McKeon et al., 1999). Koster et al. (2007) showed that the downregulation of ΔNp63 in the epidermis of newborn mice results in CHUK mRNA down-regulation as well as in severe skin fragility characterized by multiple skin erosions. This skin phenotype is similar to that observed in AEC syndrome (Marinari et al., 2008). In accordance, Marinari et al. (2008) confirmed that CHUK and p63 are co-localized in the human epidermis and the ΔNp63 isoform is required for CHUK expression in differentiating keratinocytes. Furthermore, mutant p63 protein expressed in ED patients exhibit defects in inducing CHUK expression. Consistent with these findings, we showed that CHUK and p63 co-express in several epithelial cells (Figure 3). Taken together, CHUK acts as a critical p63 downstream target gene in the regulation of epidermal morphogenesis and therefore, the variant we identified in this patient is highly likely to be causal for the EEC/AEC-like phenotypes. It will also be interesting to test the effect of this variant on the kinase activity and nuclear localization of CHUK and the CHUK-mediated negative feedback loop.
Besides the CHUK variant, two other de novo variants were identified, affecting PTGER4 and IFIT2. Neither of these genes seems to have a function in epidermal development or to be a part of the TP63 network. PTGER4 (Prostaglandin E Receptor 4) encodes a transmembrane receptor which is a member of the G-protein coupled receptor (GPCR) family. It is one of the four receptors for prostaglandin E2, which regulates multiple aspects of the immune system and may be involved in the neonatal adaptation of the circulatory system (Segi et al., 1998), osteoporosis (Yoshida et al., 2002), as well as initiation of skin immune responses (Kabashima et al., 2003). This receptor is involved in T-cell factor signaling. PTGER4 has been implicated in inflammatory bowel disease (IBD). The PTGER4 variant (g.40691969 C>T; p.Ser319Phe) identified in the index patient affects an amino acid in the TM-7 transmembrane region of the protein. Therefore, this variant could affect the function of PTGER4 and may have a role in the immune and growth-related phenotypes present in the patient. IFIT2 (Interferon-Induced protein with tetratricopeptide repeats 2) encodes for an antiviral protein which inhibits expression of viral messenger RNAs. Based on the nature of the variant, and molecular structural position and the function of the gene, the IFIT2 variant (g. 91066224 A>G; p. Asn171 Asp) identified in the index patient is unlikely to contribute to the diseases.
In conclusion, we report a patient with EEC/AEC syndrome-like phenotype with additional hypogammaglobulinemia. The heterozygous de novo variant in CHUK identified in the index patient is most likely causative for the EEC/AEC-like phenotypes. The difference in zygosity of the mutations and potentially affected pathways could explain the less severe ED phenotype and the additional immune-related phenotype in the present patient, as compared to those with previously reported Cocoon syndrome and BPS that are also caused by CHUK mutations. The PTGER4 variant could have a role, alone or in combination with the CHUK variant, in the immune and growth-related phenotypes present in the patient. Further functional analysis of this variant would be necessary to confirm its contribution to the observed phenotypes in the patient. We also suggest testing CHUK for mutations in additional patients with the combination of EEC/AEC syndrome-like and immune-related phenotypes, such as one presented here to affirm the role of CHUK further.
ACKNOWLEDGMENTS
Authors would like to thank the patient and the family involved in this study. Authors also thank Prof. Han G. Brunner for his advice on this work.
CONFLICTS OF INTEREST
Authors declare that they have no conflicts of interest.