A new family with an SLC9A6 mutation expanding the phenotypic spectrum of Christianson syndrome
Abstract
Using targeted next generation sequencing, we have identified a splicing mutation (c.526-9_526-5del) in the SLC9A6 gene in a 9-year-old boy with mild intellectual disability (ID), microcephaly, and social interaction disabilities. This intronic microdeletion leads to the skipping of exon 3 and to an in-frame deletion of 26 amino acids in the TM4 domain. It segregates with cognitive impairment or learning difficulties in other members of the family. Mutations in SLC9A6 have been reported in X-linked Christianson syndrome associating severe to profound intellectual deficiency and an Angelman-like phenotype with microcephaly, absent speech, ataxia with progressive cerebellar atrophy, ophthalmoplegia, epilepsy, and neurological regression. The proband and his maternal uncle both have an attenuated phenotype with mild ID, attention deficit disorder, speech difficulties, and mild asymptomatic cerebellar atrophy. The proband also have microcephaly. The mutation cosegregated with learning disabilities and speech difficulties in the female carriers (mother and three sisters of the proband). Detailed neuropsychological, speech, and occupational therapy investigations in the female carriers revealed impaired oral and written language acquisition, with dissociation between verbal and performance IQ. An abnormal phenotype, ranging from learning disability with predominant speech difficulties to mild intellectual deficiency, has been described previously in a large proportion of female carriers. Besides broadening the clinical spectrum of SLC9A6 gene mutations, we present an example of a monogenic origin of mild learning disability. © 2016 Wiley Periodicals, Inc.
INTRODUCTION
Next generation sequencing (NGS) of a panel of genes or of the entire exome is a very powerful tool to determine the genetic causes of these genetically heterogeneous monogenic disorders [Hennekam and Biesecker, 2012]. Since the development of these new technologies, a new approach to diagnosis of genetic diseases, called reverse phenotyping, has been developed. This consists in unraveling the genetic cause of a disease by determining what clinical phenotypes arise as a result of particular genetic sequences.
So far, 60 patients have been described in the literature with a clinical diagnosis of X-linked Christianson syndrome [Christianson et al., 1999; Gilfillan et al., 2008; Garbern et al., 2010; Schroer et al., 2010; Takahashi et al., 2011; Mignot et al., 2013; Riess et al., 2013; Bosemani et al., 2014; Pescosolido et al., 2014; Zanni et al., 2014]. The clinical features include severe to profound intellectual deficiency combined with additional manifestations such as microcephaly, absent speech, ataxia with progressive cerebellar atrophy and progression to spasticity, ophthalmoplegia, early onset seizures of variable types, and neurologic regression [Christianson et al., 1999; Schroer et al., 2010; Mignot et al., 2013; Pescosolido et al., 2014]. The molecular basis of Christianson syndrome is Solute Carrier Family 9, Member 6 (SLC9A6, also called Sodium/Hydrogen Exchanger 6; NHE6) loss of function mutation. Such genetic variations lead to a loss of function of a ubiquitously expressed Na+/H+, exchanger protein involved in neuronal development. The pathogenic mutations alter endosomal–lysosomal function, leading to a reduction in neurite outgrowth and arborization. The main consequence for brain development is cerebellar atrophy with progressive neurodegeneration and ataxia [Garbern et al., 2010; Stromme et al., 2011; Ouyang et al., 2013; Bosemani et al., 2014].
Here, we report an example of reverse phenotyping strategy in a family with SLC9A6 splice mutation identified by NGS. In this family, males had mild ID and females had language/learning disabilities thus expanding the clinical spectrum of SLC9A6 mutations.
PATIENTS AND METHODS
Patients
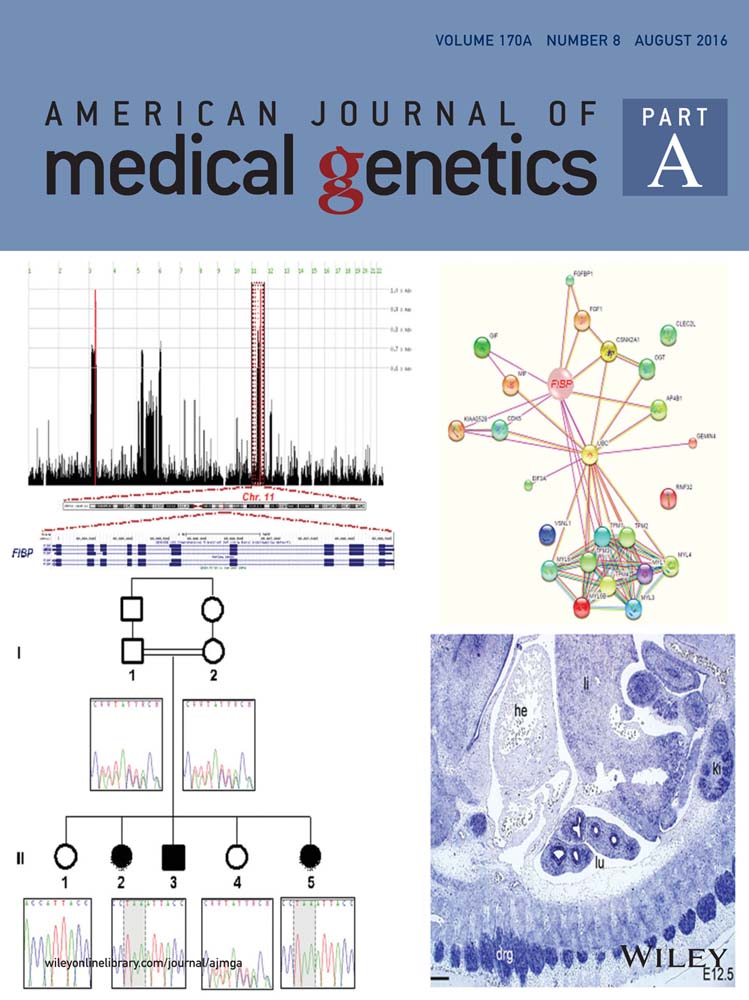
The male index case II–4 (Fig. 2A) was part of a series of 106 patients with undiagnosed ID studied by targeted-sequencing of 217 ID-genes [Redin et al., 2014]. He was born without neonatal complications at 38 weeks gestation and had weight 3140 g (50th centile); length 48.5 cm (35th centile) and occipitofrontal circumference (OFC) 32.5 cm (10th centile). Early gross motor development was normal, with walking acquired at 1 year of age. However, severe language delay was noted (only syllables at age 3 years, association of two words at age 5 years, with articulation deficiency). Mild ID was evident, and he thus attended a school for children with special needs. Only one episode of generalized tonic-clonic seizures at age 3 years was reported, first treated by Valproate. No treatment was later required, there was no recurrence of seizure after discontinuation of treatment and EEGs remained normal. Investigations included metabolic evaluations, standard karyotyping and 44 K array-CGH, cerebral CT scan, and FRAXA, ARX and PQPB1 analysis, all of which were normal. A cerebral MRI at age 5 years revealed mild cerebellar atrophy (Fig. 1C and D).
At the last examination at 9 years of age, a progressive microcephaly was noted (weight 21.6 kg [3rd centile], height 123.5 cm [5th centile], OFC 49 cm [< 3rd centile]). No ataxia, cerebellar symptoms, or pyramidal signs were noted. Facial movements were normal. There was no evidence of facial dysmorphism (Fig. 1A and B). He displayed agitation and had difficulty following instructions. Convergent strabismus without oculomotor palsy was noted. Speech had improved as he was able to utter complete sentences, but he had a persistent speech disorder.
Patient I–4 was 40 years old and was the maternal uncle of the index case. He had mild ID with an OFC at 55 cm (10th centile). Severe speech delay in childhood that required speech therapy was reported. He lived with his wife without special help and worked in a normal environment (household chores, fork-lift truck driver). He now has a normal speech. No ataxia or motor regression was noted at the clinical examination. No seizures were reported.
Patient I–2 was the index case's mother (Fig. 2A). She described having learning difficulties. She is now a house maid. Organizing family life and her children's education was difficult for her.
Patients II–1, II–3, II–5 were the index case's sisters (Fig. 2A). They all had learning disabilities, leading to difficulties at school. Motor acquisition in all three was normal, but they all displayed a constitutive speech disorder that required speech therapy. They had normal neurological examinations without microcephaly.
METHODS
Genetic Studies
Sequencing
DNA from the proband was extracted and submitted to target custom capture enrichment using probes covering the coding sequence of 217 genes known to cause ID (SureSelect, Agilent). This enriched library was sequenced in HiSeq2500 (Illumina) with a 100 bp paired-end run. Read mapping and variant calling were performed following standard procedures. VaRank, an in-house software which collects variant-specific information via Alamut version 2.2 (Interactive Biosoftware, Rouen, France) was used to rank the variants according to their predicted pathogenicity [Redin et al., 2014; Geoffroy et al., 2015]. The Exome Variant Server (EVS) and dbSNP were used to test for the presence of the variation in the general population. PCR amplification using specific primers (5′-TCT CCT GGT CGT TTG GTT GAG AAG-3 and 5′-GGA AAG AAC AGC CCA TTT CTG AAG TG-3′) and Sanger sequencing were used to confirm the presence of the mutation in the proband and in the different family members.
In silico splicing predictions
The potential effect of the variant was calculated using BDGP Splice Site Prediction by Neural Network [Reese et al., 1997], MaxEntScan [Yeo and Burge, 2004], Human Splicing Finder [Desmet et al., 2009], and GeneSplicer [Pertea et al., 2001].
RNA analysis
Total cellular RNA from the proband was isolated from blood using the PAXgene 96 Blood RNA Kit (PreAnalytix). RNA was reverse-transcribed using MMLV reverse transcriptase (Life Technologies Inc.) and complementary DNA was PCR-amplified with specific primers (SLC9A6 _exon2_F: 5′-ATG GCA TTC ATG TTC CGA GT-3′ and SLC9A6 _exon4_R: 5′-AAT GGC ACC AAA CAG TAG GC-3′). The PCR products obtained for the patient and unrelated individuals were analyzed by migration on a 2,100 Bioanalyzer instrument (Agilent Technology). The experiments were done in triplicate for the proband and one unrelated individual. The intensity of each band was calculated using 2,100 Expert Software. The different fragments obtained were subcloned into pGEM-T plasmid using pGEM-T EASY SYSTEM (Promega) and sequenced (GATC Biotech, Germany).
X-inactivation assay in female patients
This assay was performed as previously reported [Allen et al., 1992; Lau et al., 1997], with some slight modifications, notably the use of fluorescent primers and the detection mode. X-inactivation status was measured on blood DNA at the HUMARA [Allen et al., 1992] and FRAXA loci. PCR amplification of the polymorphic repeats at both loci were performed with specific FAM-labeled primers on DNA either undigested or HpaII-digested (digestion of unmethylated alleles). Amplification products were migrated on an ABI PRISM 3,500 Genetic Analyzer (Applied Biosystems, CA) to determine the area under the curve (AUC) for each allele. The X-chromosome inactivation (XCI) ratios were calculated as previously described (Lau et al. Am J Hum Genet 1997): XCI = (AUCallele1 − HpaII/AUCallele1 − undigested)/[(AUCallele1 − HpaII/AUCallele1 − undigested) + (AUCallele2 − HpaII/AUCallele2 − undigested)]. X-inactivation was considered as biased when it was greater than or equal to 85%.
Multidisciplinary speech evaluation
Multidisciplinary evaluation was done in the two male patients and in all four female carriers. The methods used are summarized in Tables I and II.
| Patient II–4 (9 years) | Patient I–4 (40 years) | |
|---|---|---|
| Neuropsychological evaluation | Mild intellectual disability (Raven matrices) Language: age equivalent 3 years and 4 months. Social interactions, activities related to acquisition of autonomy and motor function: Age equivalent 5 ½ years, 6 years (Vineland) | Mild intellectual disabilities (WAIS-IV) VCI = 61 PRI = 72 WMI = 66 PSI = 75 FSIQ = 60 |
| Verbal langage | Oral language disorders affecting the receptive and expressive language. Short sentences, impaired articulation | Normal sentences, no evaluation |
| Psychiatric evaluation | Absence of stereotypical behavior, restricted communication and creativity, included in intellectual deficiency. Good quality reciprocal social interactions, but hindered by impaired oral communication. No elements in favor of autism spectrum disorders (ASD) | No evaluation |
| No signs of anxiety |
- FSIQ, Full Scale IQ score; PRI, Perceptual Reasoning Index; VCI, Verbal Comprehension Index; WMI, Working Memory Index.
| Patient II–1 | Patient II–3 | Patient II–5 | Patient I–2 | |
|---|---|---|---|---|
| Age (years) | 13 | 12 | 6.5 | 42 |
| OFC | − 0.5 SD | −1 SD | Mean | Mean |
| Neuropsychological evaluation | VCI = 57 PRI = 90 WMI = 76 PSI = 83 (WISC IV) Joint attention deficit | VCI = 53 PRI = 88 PSI = 96 WMI = 91 (WISC IV) | VIQ = 66 PIQ = 82 PSQ = 89 (WPPSI) Bimodal joint attention deficit | VCI = 75 PRI = 87 WMI = 82 PSI = 94 (WAIS IV) |
| Speech evaluation | Dyslexia/dysorthographia | Dyslexia/dysorthographia, limited vocabulary | Sequelae of language disorders (limited vocabulary, errors of syntax, absence of chronological narrative) | Not performed |
| Occupational therapy | Disorders in writing subsequent to impaired oral language Neurovisual disorders (immaturity of the ocular motor saccades) with repercussions on visuo-motor abilities Disorders with single-hand motor function (right and left) | Major neurovisual disorders (immaturity of the ocular motor saccades), writing disorders directly related to the neurovisual disorders and impaired oral and written language | Neurovisual disorders (immaturity of the ocular motor saccades); visuospatial disorders (no mental representation of objects and the environment). Disorders that hindered writing and precision | Not performed |
| Psychiatric evaluation | Excessive inhibition, with avoidance of situations that involve spontaneous language | Psychoaffective inhibition, with avoidance of situations that involve spontaneous language | No psychiatric manifestation at this age | Not performed |
| Conclusion | Specific learning disorders of the dysphasia and dyslexia/dysorthographia type + neuro-visual disorders | Specific learning disorders of the oral and written language type related to dysphasia and dyslexia/dysorthographia | Oral language disorders of the dysphasia type + neurovisual and visuospatial disorders | Specific learning disorders |
- FSIQ, Full Scale IQ score; PIQ, Performance Index Quotient; PSI, Processing speed Index; PSQ, Processing Speed Quotient; PRI, Perceptual Reasoning Index; VCI, Verbal Comprehension Index; VIQ, Verbal Index Quotient; WMI, Working Memory Index.
RESULTS
Genetic Studies
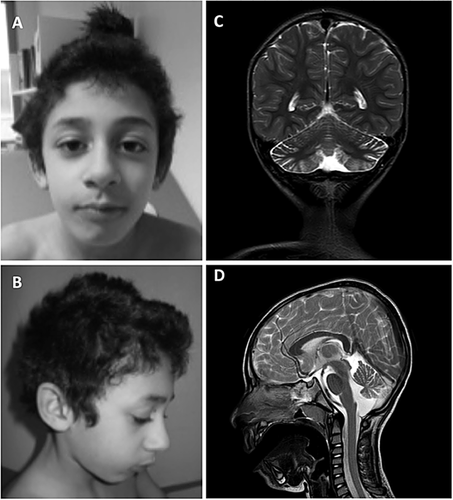
A splicing mutation (c.526-9_526-5del) in the SLC9A6 gene was identified by targeted NGS in the patient II–4 [Redin et al., 2014]. It segregates with cognitive impairment or learning difficulties in the other family members (Fig. 2A). The deletion of five nucleotides affects the prediction scores for using the splice acceptor site (SAS) located at the end of intron 2, according to MaxEnt Splice, Nnsplice, GeneSplicer, and HSF prediction programs (Fig. 2B). Blood RNA analysis revealed that different splicing events occur between exons 2 and 4 with the wild-type allele and with the c.526-9_526-5del mutation (Fig. 2C). In normal blood, more than 90% of the transcripts include the entire exon 2 (form 1, coding for the longest isoform NM_001042537) and less than 10% comprised only a short exon 2, using an alternative splice donor site located 96 nucleotides upstream (form 1s, coding for the shortest isoform NM_006359) (Fig. 2D). In the proband, four different transcripts were identified: transcripts where exon 3 was included (form 1 and form 1s) and transcripts where exon 3 was skipped, with either the entire exon 2 (form 2) or the short exon 2 (form 2s) (Fig. 2C). The skipping of exon 3 occurs in the majority of the transcripts (almost 90%, Fig. 2D). The skipping of this exon, caused by the intronic deletion c.526-9_526-5del, is predicted to lead to an inframe deletion of 26 amino acids (p.Val177_Arg202del) in the fourth transmembrane domain (TM4) (Fig. 2E) and might therefore affect the folding of the protein in the membrane.
Regarding X-inactivation studies, the mother I–2 and the sister II–3 had unbiased X-inactivation studies. II–5 had a biaised X-inactivation at the HUMARA and FRAXA loci (84/16%).
Multidisciplinary Evaluation
Neuropsychological evaluations of affected males indicated the presence of mild ID; the results are summarized in Table I Multidisciplinary neuropsychological, speech, and occupational therapy evaluations of the female carriers showed dissociation between the performance intelligence quotient (PIQ) and verbal intelligence quotient (VIQ) (summarized in Table II). These results are suggestive of probable dysphasia with dyslexia.
DISCUSSION
We report on a family with a novel splicing SLC9A6 variant, expanding the clinical spectrum of Christianson syndrome. The two males in this family only displayed mild ID with speech difficulties associated with microcephaly and asymptomatic cerebellar atrophy in the proband. Reviewing the 60 cases of Christianson syndrome described in the literature, the patients described here were much less severely affected than previously described patients [Christianson et al., 1999; Gilfillan et al., 2008; Garbern et al., 2010; Schroer et al., 2010; Takahashi et al., 2011; Mignot et al., 2013; Riess et al., 2013; Bosemani et al., 2014; Pescosolido et al., 2014; Zanni et al., 2014]. Indeed, all patients have profound ID, except for one who had moderate ID [Schroer et al., 2010]. In the largest series of 14 patients recently reported by Pescosolido et al. [2014], all males were described as nonverbal and had ID, epilepsy, and ataxia. Many had prior diagnoses of autism and/or Angelman syndrome. Other neurologic symptoms included eye movement abnormalities (79%), postnatal microcephaly (92%), and magnetic resonance imaging evidence of cerebellar atrophy (33%). Regression was noted in 50%, with recurrences of loss of speech and/or inability to walk. Medical symptoms, particularly gastrointestinal symptoms (feeding difficulties, gastroesophageal reflux), were common. Height and body mass index measures were below normal ranges in most participants. Behavioral symptoms included hyperkinetic behavior (100%), and a majority exhibited a high pain threshold.
In our family, the epileptic manifestations were absent in I–4 and mild and transitory in patient II–4, who requires no treatment today. This contrasts with seizures that are often described in the literature as resistant to multiple therapies. In addition, no motor difficulties, ataxia, spasticity, or motor regression were noted, even in our 40-year-old patient. Both patients had severe speech difficulties during infancy. However, the older patient had normal language in adulthood. Evaluation of our index case showed that he began to express himself with short sentences at age 7 years. Nearly all patients described in the literature had no language except for two patients reported by Garbern et al. [2010]. However, their level of language was not described and they were reported to have profound ID. We searched for genotype-phenotype correlations in order to explain the milder phenotype in our patients. Exon 3 skipping had been already described in three families with a severe phenotype [Gilfillan et al., 2008; Pescosolido et al., 2014]. The milder phenotype in our family might be explained by the remaining 10% of the normal transcript. However, our patients’ follow up will continue since late regression (40–60 year-old) has already been described in association with SLC9A6 mutations [Garbern et al., 2010].
We were able to perform detailed evaluations in the four female carriers of the family. They all displayed a clear dissociation between VIQ and PIQ, with dysphasia, dyslexia, writing difficulties (dysorthographism), and visual disorders. Manifestations in females are less frequently described than those in males. We found reports of 27 female carriers with available clinical descriptions. Fifteen were reported to have learning difficulties or mild to moderate ID, whereas 12 had normal development with no detailed evaluations (Table III). When described or evaluated, a VIQ inferior to the PIQ and speech deficits were commonly noted [Schroer et al., 2010]. The high prevalence of cognitive manifestations in female carriers strongly argues for a carrier phenotype, and SLC9A6 mutations in females can therefore be a monogenic cause of constitutive speech disorders.
| Present report | Christianson et al. [1999] | Takahashi et al. [2011] | Gilfillan et al. [2008] | Garbern et al. [2010] | Schroer et al. [2010] family 1 | Riess et al. [2013] | Zanni et al. [2014] | Pescosolido et al. [2014] | Total | |
|---|---|---|---|---|---|---|---|---|---|---|
| Number of female carriers with available data | 4 | 10 | 1 | 4 | 1 | 3 | 1 | 1 | 2 | 27 |
| Existence of learning disabilities | 4/4 Dysphasia | 3/10 | – | 1/4 severe dyslexia | Mild ID | 3/3 1 Learning deficit and speech disorder 1 school difficulties 1 mild ID with hyperkinesis and mild truncal ataxia | – | Mild ID | 1 moderate ID, speech and language delay, childhood onset schizophrenia and autism spectrum disorder | 15 |
| 2 Learning difficulties, 1 mild ID | ||||||||||
| 1 mild ID with microcephaly | ||||||||||
| Normal development | – | 7/10 | 1 | 3/4 | – | – | 1 | – | – | 12 |
In conclusion, by sequencing panels of genes in patients with no precise clinical diagnosis, NGS can broaden the clinical variability associated with a known gene. We also argue that SLC9A6 gene mutations in females could be responsible for a monogenic cause of mild learning disability/constitutive speech disorders.
ACKNOWLEDGMENTS
The authors thank the family for their participation in the study. The authors also thank the Regional Council of Burgundy, the Fondation Jerome Lejeune, and the Agence de Biomedecine for their financial support. CR is a fellowship student from the APLM and Groupement d'Intérêt Economique-Centre Européen de Recherche en Biologie et en Médecine.