SMARCE1, a rare cause of Coffin–Siris Syndrome: Clinical description of three additional cases
Abstract
Coffin–Siris syndrome (CSS, MIM 135900), is a well-described, multiple congenital anomaly syndrome characterized by coarse facial features, hypertrichosis, sparse scalp hair, and hypo/aplastic digital nails and phalanges, typically of the 5th digits. Mutations in the BAF (SWI/SNF)-complex subunits (SMARCA4, SMARCE1, SMARCB1, SMARCA2, ARID1B, and ARID1A) have been shown to cause not only CSS, but also related disorders including Nicolaides–Baraitser (MIM 601358) syndrome and ARID1B-intellectual disability syndrome (MIM 614562). At least 200 individuals with CSS have been found to have a mutation in the BAF pathway. However, to date, only three individuals with CSS have been reported to have pathogenic variants in SMARCE1. We report here three additional individuals with clinical features consistent with CSS and alterations in SMARCE1, one of which is novel. The probands all exhibited dysmorphic facial features, moderate developmental and cognitive delay, poor growth, and hypoplastic digital nails/phalanges, including digits not typically affected in the other genes associated with CSS. Two of the three probands had a variety of different organ system anomalies, including cardiac disease, genitourinary abnormalities, feeding difficulties, and vision abnormalities. The 3rd proband has not had further investigative studies. Although an increasing number of individuals are being diagnosed with disorders in the BAF pathway, SMARCE1 is the least common of these genes. This report doubles the number of probands with these mutations, and allows for better phenotypic information of this rare syndrome. © 2016 Wiley Periodicals, Inc.
INTRODUCTION
Coffin–Siris Syndrome (CSS, MIM 135900) is a now well-described, multiple congenital anomaly syndrome, found to be caused by mutations in the BRG- or HRBM- associated factor (BAF) pathway in humans. Within this family of genes, pathogenic variants in ARID1B (the most common), ARID1A, SMARCB1, SMARCA4, and SMARCE1 have all been found in individuals with clinical phenotypes consistent with CSS. More recently, pathogenic alterations of the SOX11 gene were also found in probands with neurodevelopmental delay and features of CSS [Hempel et al., 2016]. In general, patients with pathogenic variants in all the BAF pathway genes tend to share the core features of some degree of developmental and growth delay, hypo/aplasia of the nails or phalanges, typically of the 5th digits, dysmorphic facial features, and either hypertrichosis or hirsutism with paradoxically sparse scalp hair. They may also have a number of different organ-system anomalies including feeding difficulties, cardiac abnormalities, and delayed growth and development [Coffin and Siris, 1970; Fleck et al., 2001; Schrier et al., 2012; Kosho et al., 2013; Santen et al., 2013; Wieczorek et al., 2013].
Recently, genotype–phenotype correlations in CSS have begun to emerge. Individuals with SMARCB1 pathogenic variants, for example, tend to be described as having the most “coarse” facial features. They also have structural central nervous system (CNS) abnormalities and tend to be severely developmentally delayed. Individuals with SMARCA4 pathogenic variants, on the other hand, do not have facial features that are as coarse, and have a broader range of developmental delays, with some individuals described as walking independently and speaking well [Kosho et al., 2014].
To date, only three individuals with SMARCE1 pathogenic variants have been reported [Tsurusaki et al., 2012, 2014; Kosho et al., 2013; Santen et al., 2013; Wieczorek et al., 2013]. These individuals have been described as having mild to moderate growth impairment and all had feeding difficulties, with moderate to severe cognitive and developmental delay. They were also noted to have structural CNS abnormalities, congenital heart disease, and visual impairment. Physically, they displayed thick eyebrows, a thick lower lip but thin upper lip, hypertrichosis, and all had hypoplastic nails or phalanges to the digits.
Here, we add to the literature three additional individuals with features of CSS and pathogenic variants in SMARCE1. They all displayed the typical hypo/aplastic digital phalanges or nails to the fingers or toes, including digits which are not usually affected in the CSS caused by other gene mutations. They also share coarse facial features, ectodermal changes, and varying organ system anomalies, including significant feeding difficulties and delayed growth and development. In adding these probands to the literature, this report broadens the currently understood phenotype of this rare mutation in this increasingly recognized syndrome.
MATERIALS AND METHODS
All three individuals have been registered in the Coffin–Siris and BRG1- or HRBM- Associated Factor Related Disorders (CSS-BAF) Registry.
Patients 1 and 3
Exome sequencing (ES) was performed on exon targets isolated by capture using the Agilent SureSelect Human All Exon version 5 kit (Agilent Technologies, Santa Clara, CA) following standard procedures described elsewhere [Bhoj et al., 2015; Miyake et al., 2015]. The prepared sample was sequenced on HiSeq2500 according to the manufacturer's recommendations (Illumina, San Diego, CA). The reads were mapped to human reference genome hg19 by Novoalign. The variant call and annotation were performed with Genome Analysis Toolkit and ANNOVAR. For patient 3, only the patient's sample was applied for ES while parental samples were analyzed in patient 1 for Sanger confirmation. The candidate variants in known genes for CSS were validated by Sanger sequencing to occur as de novo.
Patient 2
Complete sequence analysis of the coding regions of the following genes were performed: ARID1A, ARID1B, PHF6, SMARCA2, SMARCA4, SMARCB1, and SMARCE1. Targets of interest were enriched and amplified using the Agilent SureSelect System (Agilent Technologies). DNA was sequenced using Illumina technology through the University of Chicago (Illumina).
CLINICAL REPORTS
Specific clinical details and are outlined in Table I.
| Features | Patient 1 | Patient 2 | Patient 3 | Tsurusaki et al. [2012] and Kosho et al. [2013] | Wieczorek et al. [2013] | Santen et al. [2013] | Total |
|---|---|---|---|---|---|---|---|
| SMARCE1 variant | p.Tyr126Asp | p.Tyr73Cys | p.Arg105Gln | p.Tyr73Cys | p.Tyr73Ser | p.Arg105Gln | |
| Origin | de novo | Unknown | de novo | de novo | de novo | Inherited from affected mother | |
| Gender | Male | Female | Female | Female | Female | Male | |
| Age, years | 2 | 1 | 8 | 17 | 3 | 19 | |
| Gestation | 37 weeks | 37 5/7 weeks | 36 weeks | 37 weeks | 38 weeks | Not reported | |
| BW (Kg) | 3.06 Kg | 1.7 Kg | 1.8 Kg | 1.64 Kg | 2.2 | Not reported | |
| Microcephaly | (−) | (+) | (+) | (+) | (+) | (−) | 4/6 |
| HEENT | Cleft palate, laryngotracheomalacia | High palate, retromicrognathia | High palate | Cleft palate | Suspected cleft palate | (−) | 5/6 |
| Hypotonia | (+) | hypertonia | (−) | (+) | (−) | (−) | 2/6 |
| Seizures | (−) | (−) | (−) | (+) | (−) | (+) | 2/6 |
| Cardiac | MS, LVH, left ventricular diastolic dysfunction | PDA, ASD | Not reported | AS, TS, MS, CAA | Dex, ASD, PH | (−) | 4/5 |
| GI | Feeding difficulties | Feeding difficulties, G-tube | (−) | Feeding difficulties, G-tube | Feeding difficulties, G-tube | Feeding difficulties, pyloric stenosis | 5/6 |
| GU | Cryptorchidism | (−) | (−) | Not reported | Not reported | (−) | 1/4 |
| Developmental delay | (+) 19 months, non-verbal, crawling | (+) 1 year, rolls over | (+) 8 years, DQ 45 | (+) 14 years, non-verbal, walking | (+) sat at 18 months, walked at 54 months | (+) Moderate DD | 6/6 |
| Structural brain | White matter volume loss, thin optic nerve/chiasm | Not reported | Not reported | CH | CH, DW, ACC | None reported | 3/4 |
| Visual anomalies | Decreased visual acuity | Nystagmus | (−) | None reported | None reported | Myopia | 3/6 |
| Hearing loss | (−) | (+) | (−) | (+) | None reported | (−) | 2/6 |
| Recurrent infections | (−) | (+) | (−) | (+) | (+) | (+) | 4/6 |
| Ortho | (−) | 13 ribs bilaterally | (−) | Scoliosis | Not reported | Pectus excavatum, dislocated elbows | 3/6 |
| Digital anomalies | Hypoplastic 5th toenails, midline split of right hallux toenail | Absent nails on great toes | Hypoplastic second digit nails bilaterally | Hypoplasic 5th digit nails fingers/toes | Hypo/aplastic 5th fingers/toes and nails | Hypo/aplastic 5th fingers and nails | 6/6 |
| Dental anomalies | (+) | Not reported | (−) | (+) | Not reported | (−) | 2/6 |
| Facial features | |||||||
| Coarse appearance | (+) | (+) | (+) | (+) | (+) | (+) | 6/6 |
| Flat nasal bridge | (−) | (+) | (−) | (+) | (+) | (+) | 4/6 |
| Wide mouth | (+) | (+) | (+) | (+) | (+) | (−) | 5/6 |
| Thick lips | (+) | (+) | (+) | (+) | (+) | (+) | 6/6 |
| Thick eyebrows | (+) | (−) | (+) | (+) | (+) | (+) | 5/6 |
| Abnormal ears | (−) | (+) | (+) | (+) | (+) | (+) | 5/6 |
| Sparse scalp hair | (+) | (+) | (+) | (+) | (+) | (−) | 5/6 |
- ACC, Absent corpus collosum; AS, Aortic stenosis; ASD, Atrial septal defect; BW, Birth weight; CH, Congenital hydrocephalus; CAA, Coronary artery anomaly; DW, Dandy walker malformation; DQ, Developmental quotient; DD, Developmental delay; Dex, Dextrocardia; FOC, Fronto-occipital circumference; GI, Gastrointestinal; G-Tube, Gastric tube; GU, Genitourinary; HEENT, Head, eyes, ears, nose, and throat; Kg, Kilograms; LVH, Left ventricular hypertrophy; MS, Mitral stenosis; PDA, Patent ductus arteriosus; PH, Pulmonary hypertension; TS, Tricuspid stenosis; +, Feature present; −, Feature absent.
Patient 1
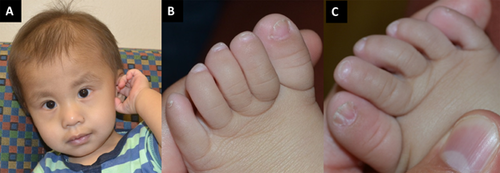
The male proband was born at term weighing 3.06 kg (27th centile) to a non-consanguineous couple following an uncomplicated pregnancy and delivery. Postnatal exam revealed a heart murmur, a cleft soft palate, and cryptorchidism. Kidney and spine imaging revealed no abnormalities. Significant feeding issues were appreciated during the following months and the patient was diagnosed with laryngotracheomalacia. Developmentally, he was found to have significant hypotonia and global developmental delay. By 19 months of age, he had no words, he was crawling but not walking independently, and still had a rake-grasp. At that time his length was 78.74 cm (8th centile), weight was 8.75 kg (2nd centile), and head circumference was 46.9 cm (37th centile). On physical exam he was noted to have dental decay, hypoplastic 5th toenails, and a midline split on the right hallux toenail (Fig. 1). Family history was negative for orofacial clefting, developmental delay, or ectodermal-like changes. Prior testing included a normal SNP microarray.
Patient 2
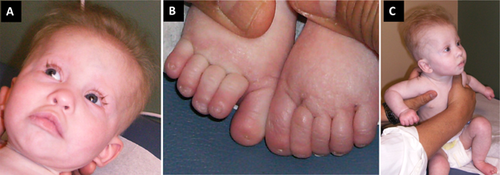
The Proband was the product of a 37 and 5/7 week gestation born after prenatal ultrasounds suggestive of intrauterine growth retardation with a suspected atrioventricular canal defect. Birth weight was 1.7 kg (<3rd centile), length 42.5 cm (3rd centile), and head circumference 29.5 cm (0.5th centile). Postnatal findings included a large patent ductus arteriosus, moderate secundum atrial septal defect, 13 ribs bilaterally, absent nails on great toes, high arched palate with retromicrognathia, and a sacral dimple/hemangioma. Head, renal, and spinal ultrasounds all were normal. As she grew, she was noted to have sparse scalp hair, nystagmus, and abnormally shaped dentition (Fig. 2). Feeding issues necessitated a feeding tube. She rolled over at 11 months of age and is now just over 1-year old as of this report. She is reported to have bilateral sensorineural hearing loss. At an evaluation at 10 months of age, weight was 4.49 kg (50th centile for 2-month old), height measured 59.8 cm (50th centile for a 2–3 month old) and head circumference was 39.4 cm (50th centile for a 3-month old). Family history was negative birth defects, developmental delay, or ectodermal-like changes. A SNP microarray returned normal.
Patient 3
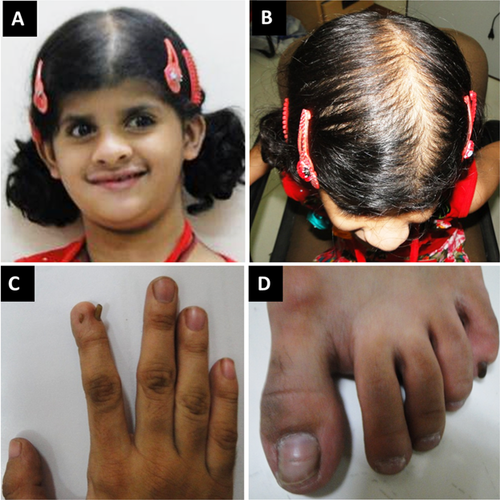
The proband was the product of a 36 week gestation born to her non-consanguineous parents weighing 1.8 kg (50th centile for a 33 week gestation). There was no history of feeding difficulties. She was reported to be in good health but her parents began to note developmental delays at a year of age. She sat unsupported at 9 months of age, stood without support at 2 years and 4 months, and walked at 2.5 years of age. She was able to say two-syllable words at 2 years. She was evaluated at age 8 years in the clinic for the delayed development and hyperactivity. She was described as having mild hirsutism, and had a hypoplastic right second digit nail (Fig. 3), along with long, slender fingers. A bone age X-ray at age 8 years showed tapering of the distal phalanges. Weight at the time measured 24 kg (50th centile), height was 132 cm (75th centile), and head circumference was 49.5 cm (25th centile). Family history was negative for birth defects, developmental delay, or ectodermal-like changes. A SNP microarray and karyotype were both normal.
RESULTS
Patient 1
After standard multiple filtering steps describe elsewhere and followed by Sanger sequencing with both parents, we identified a de novo novel substitution (c.376T > G, p.(Y126D)) in SMARCE1 (NM_003079.4) as the most likely disease-causing candidate. This variant is a non-conservative amino acid substitution (Grantham score: 160) of a polar neutral Tyrosine with a negatively charged Aspartic acid at a residue that is highly conserved across species and is located in the high mobility domain (HMG box) of the protein that had three earlier reported SMARCE1 variants in CSS. The c.376T > G variant was predicted damaging by SIFT (0.01), PolyPhen-2 (0.998), and MutationTaster. The analysis of public databases (1,000 Genomes Project, 6503 exomes from the Exome Sequencing Project (ESP6500SI), and Exome Aggregation Consortium dataset (ExAC v0.3)) and existing sequencing data from more than 2,000 ES samples in the local database did not detect any occurrence of the variant.
Patient 2
DNA sequence analysis of the SMARCE1 gene demonstrated a sequence change (c.218A > G, p.(Y73C)) in exon 5. The Y73C change affects a highly conserved amino acid residue located in a conserved protein domain that has previously been described in a least one patient with CSS in the de-novo state [Tsurusaki et al., 2012]. Parental studies were not conducted but clinically, neither parent had findings to suggest an even milder form of CSS.
Patient 3
After standard multiple filtering steps describe elsewhere and followed by Sanger sequencing with both parents, we identified a de novo substitution of the SMARCE1 gene, c.314G > A; p.R105Q (NM_003079.4) in exon 5. While the variant had conflicting in silico predictions (SIFT: tolerated, 0.13; PolyPhen-2: benign, 0.279, MutationTaster: disease-causing), the same change was previously reported in a patient with CSS who inherited the variant from his affected mother [Santen et al., 2013]. The analysis of public databases (1,000 Genomes Project, 6503 exomes from the Exome Sequencing Project (ESP6500SI), and Exome Aggregation Consortium dataset (ExAC v0.3) and existing sequencing data from more than 2,000 ES samples in the local database did not detect any occurrence of the variant.
DISCUSSION
While phenotypes of the additional BAF pathway genes all have some overlap, genotype–phenotype correlations are beginning to emerge more precisely as additional individuals are reported. In general, probands with SMARCB1 mutations appear to have the coarsest facial features and significant developmental and growth delays [Kosho et al., 2014]. Alterations in ARID1B are the most common cause of CSS, and individuals can have a range of severity in terms of physical anomalies and intellectual deficits [Kosho et al., 2014; Santen et al., 2014]. Meanwhile, probands with mutations in ARID1A tend to have the most severe phenotypes, with absent speech and intellectual disability, and severe physical complications [Kosho et al., 2014]. We describe three additional patients with alterations in the SMARCE1 gene, the rarest of those seen in individuals with a molecular diagnosis of CSS.
Overall, the three patients share a similar phenotype with the SMARCE1 probands previously reported: the probands in this report display moderate-severe intellectual and developmental delay, anomalies in a variety of organ systems, and the “classic” physical features of CSS including sparse scalp hair, coarse facial features, and digital phalanx/nail hypo/aplasia (Table I). Patient 3 displayed hypoplasia to digits other than the 5th, a finding which has been reported in patients with CSS and alteration in the BAF pathway genes, including SMARCE1 probands [Kosho et al., 2014]. Similarly to what is seen in patients with ARID1B pathogenic variants, the compilation of the only six individuals described with SMARCE1 alterations suggests that the facial features while consistently coarse, they are less severe when compared to patients with other BAF pathway pathogenic alterations [Tsurusaki et al., 2012, 2014; Kosho et al., 2013; Santen et al., 2013; Wieczorek et al., 2013]. Additionally, the earlier comparison of this limited amount of patients with SMARCE1 alterations against other patients with CSS suggests as high a frequency of growth delays and feeding difficulties, and potentially higher frequency of moderate-severe developmental and cognitive delays (Table I).
Recognizing that genotype–phenotype correlations for SMARCE1 alterations are vastly limited by the small number of patients, clinicians should continue to consider the diagnosis of CSS with patients who have 5th digit phalanx and nail anomalies, and in individuals who also have sparse scalp hair and hirsutism. SMARCE1 mutations may be more likely to be present in patients whose facial features are more variable and not as coarse as in the other BAF pathway genes. As more individuals with CSS and SMARCE1 pathogenic variants are identified, an even broader phenotype is likely to emerge and add detail to the growing spectrum of this syndrome.